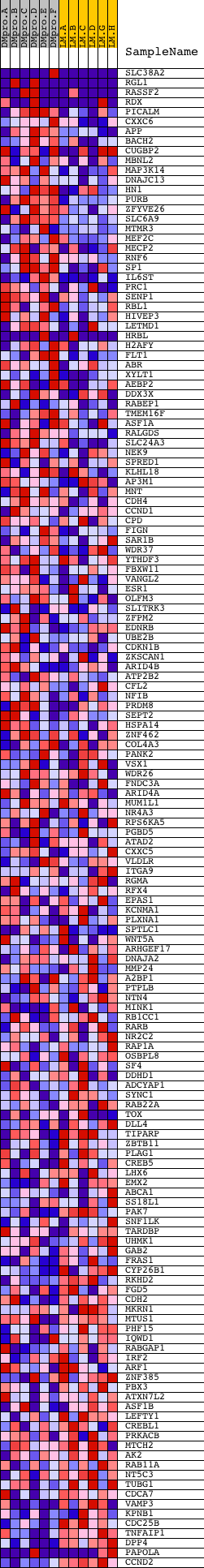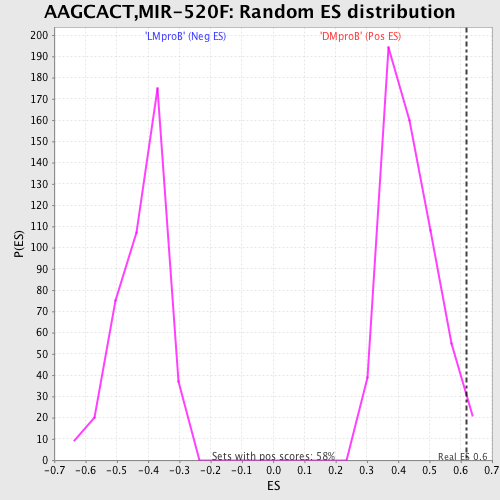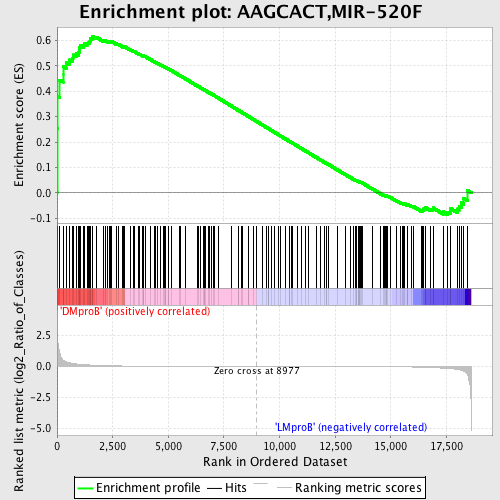
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | DMproB |
| GeneSet | AAGCACT,MIR-520F |
| Enrichment Score (ES) | 0.6174883 |
| Normalized Enrichment Score (NES) | 1.4148597 |
| Nominal p-value | 0.022530328 |
| FDR q-value | 0.8649112 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC38A2 | 22149 2219 | 4 | 4.197 | 0.2550 | Yes | ||
| 2 | RGL1 | 9723 4072 | 32 | 2.063 | 0.3789 | Yes | ||
| 3 | RASSF2 | 2802 14422 | 120 | 1.156 | 0.4445 | Yes | ||
| 4 | RDX | 9712 5371 | 268 | 0.508 | 0.4674 | Yes | ||
| 5 | PICALM | 18191 | 270 | 0.504 | 0.4980 | Yes | ||
| 6 | CXXC6 | 19747 | 405 | 0.366 | 0.5130 | Yes | ||
| 7 | APP | 4402 | 540 | 0.293 | 0.5235 | Yes | ||
| 8 | BACH2 | 8648 | 676 | 0.247 | 0.5312 | Yes | ||
| 9 | CUGBP2 | 4687 8923 | 716 | 0.234 | 0.5433 | Yes | ||
| 10 | MBNL2 | 21932 | 859 | 0.199 | 0.5477 | Yes | ||
| 11 | MAP3K14 | 11998 | 953 | 0.183 | 0.5538 | Yes | ||
| 12 | DNAJC13 | 6177 | 990 | 0.176 | 0.5626 | Yes | ||
| 13 | HN1 | 9101 | 1012 | 0.173 | 0.5719 | Yes | ||
| 14 | PURB | 5335 | 1033 | 0.170 | 0.5812 | Yes | ||
| 15 | ZFYVE26 | 9999 | 1197 | 0.146 | 0.5812 | Yes | ||
| 16 | SLC6A9 | 4783 | 1247 | 0.140 | 0.5870 | Yes | ||
| 17 | MTMR3 | 7908 13193 | 1346 | 0.126 | 0.5894 | Yes | ||
| 18 | MEF2C | 3204 9378 | 1398 | 0.121 | 0.5940 | Yes | ||
| 19 | MECP2 | 5088 | 1474 | 0.113 | 0.5968 | Yes | ||
| 20 | RNF6 | 13168 | 1486 | 0.112 | 0.6030 | Yes | ||
| 21 | SP1 | 9852 | 1489 | 0.112 | 0.6097 | Yes | ||
| 22 | IL6ST | 4920 | 1572 | 0.104 | 0.6115 | Yes | ||
| 23 | PRC1 | 12862 18203 | 1579 | 0.103 | 0.6175 | Yes | ||
| 24 | SENP1 | 10301 | 1778 | 0.087 | 0.6121 | No | ||
| 25 | RBL1 | 14373 | 2098 | 0.065 | 0.5988 | No | ||
| 26 | HIVEP3 | 4973 | 2153 | 0.063 | 0.5997 | No | ||
| 27 | LETMD1 | 22357 | 2257 | 0.058 | 0.5976 | No | ||
| 28 | HRBL | 3452 16326 3614 | 2365 | 0.053 | 0.5950 | No | ||
| 29 | H2AFY | 21447 | 2397 | 0.051 | 0.5965 | No | ||
| 30 | FLT1 | 3483 16287 | 2465 | 0.049 | 0.5958 | No | ||
| 31 | ABR | 8468 20346 1476 1429 | 2651 | 0.041 | 0.5883 | No | ||
| 32 | XYLT1 | 18113 | 2740 | 0.038 | 0.5858 | No | ||
| 33 | AEBP2 | 4359 | 2958 | 0.032 | 0.5761 | No | ||
| 34 | DDX3X | 24379 | 2969 | 0.032 | 0.5775 | No | ||
| 35 | RABEP1 | 12016 1344 | 3028 | 0.031 | 0.5762 | No | ||
| 36 | TMEM16F | 22377 | 3300 | 0.026 | 0.5631 | No | ||
| 37 | ASF1A | 20025 | 3435 | 0.023 | 0.5573 | No | ||
| 38 | RALGDS | 15066 | 3475 | 0.023 | 0.5565 | No | ||
| 39 | SLC24A3 | 14818 | 3671 | 0.020 | 0.5472 | No | ||
| 40 | NEK9 | 10141 | 3704 | 0.019 | 0.5466 | No | ||
| 41 | SPRED1 | 4331 | 3849 | 0.018 | 0.5399 | No | ||
| 42 | KLHL18 | 11291 | 3884 | 0.017 | 0.5391 | No | ||
| 43 | AP3M1 | 7059 | 3891 | 0.017 | 0.5398 | No | ||
| 44 | MNT | 20784 | 3972 | 0.016 | 0.5365 | No | ||
| 45 | CDH4 | 8728 | 4204 | 0.014 | 0.5249 | No | ||
| 46 | CCND1 | 4487 4488 8707 17535 | 4387 | 0.013 | 0.5158 | No | ||
| 47 | CPD | 20344 4556 | 4410 | 0.013 | 0.5154 | No | ||
| 48 | FIGN | 14575 | 4528 | 0.012 | 0.5098 | No | ||
| 49 | SAR1B | 20891 | 4663 | 0.011 | 0.5033 | No | ||
| 50 | WDR37 | 9874 5502 | 4778 | 0.011 | 0.4977 | No | ||
| 51 | YTHDF3 | 15630 10469 6019 | 4838 | 0.010 | 0.4952 | No | ||
| 52 | FBXW11 | 20926 | 4875 | 0.010 | 0.4939 | No | ||
| 53 | VANGL2 | 4109 8220 | 4882 | 0.010 | 0.4942 | No | ||
| 54 | ESR1 | 20097 4685 | 5004 | 0.010 | 0.4882 | No | ||
| 55 | OLFM3 | 1864 1769 15441 | 5027 | 0.010 | 0.4876 | No | ||
| 56 | SLITRK3 | 11816 | 5144 | 0.009 | 0.4819 | No | ||
| 57 | ZFPM2 | 22483 | 5490 | 0.008 | 0.4637 | No | ||
| 58 | EDNRB | 21728 3564 | 5544 | 0.008 | 0.4613 | No | ||
| 59 | UBE2B | 10245 | 5759 | 0.007 | 0.4501 | No | ||
| 60 | CDKN1B | 4512 8730 | 6331 | 0.005 | 0.4195 | No | ||
| 61 | ZKSCAN1 | 7927 | 6343 | 0.005 | 0.4192 | No | ||
| 62 | ARID4B | 21705 3233 | 6436 | 0.005 | 0.4145 | No | ||
| 63 | ATP2B2 | 17038 | 6578 | 0.005 | 0.4072 | No | ||
| 64 | CFL2 | 2155 21069 | 6622 | 0.005 | 0.4051 | No | ||
| 65 | NFIB | 15855 | 6623 | 0.005 | 0.4054 | No | ||
| 66 | PRDM8 | 8084 | 6651 | 0.004 | 0.4042 | No | ||
| 67 | SEPT2 | 5159 9453 | 6787 | 0.004 | 0.3971 | No | ||
| 68 | HSPA14 | 14695 | 6830 | 0.004 | 0.3951 | No | ||
| 69 | ZNF462 | 6321 | 6930 | 0.004 | 0.3900 | No | ||
| 70 | COL4A3 | 8766 | 7015 | 0.004 | 0.3856 | No | ||
| 71 | PANK2 | 14838 | 7071 | 0.004 | 0.3829 | No | ||
| 72 | VSX1 | 2766 14398 | 7243 | 0.003 | 0.3738 | No | ||
| 73 | WDR26 | 13733 | 7824 | 0.002 | 0.3425 | No | ||
| 74 | FNDC3A | 6672 21756 | 8149 | 0.001 | 0.3251 | No | ||
| 75 | ARID4A | 2080 6215 | 8168 | 0.001 | 0.3242 | No | ||
| 76 | MUM1L1 | 10884 | 8271 | 0.001 | 0.3187 | No | ||
| 77 | NR4A3 | 9473 16212 5183 | 8351 | 0.001 | 0.3145 | No | ||
| 78 | RPS6KA5 | 2076 21005 | 8609 | 0.001 | 0.3006 | No | ||
| 79 | PGBD5 | 9960 | 8617 | 0.001 | 0.3003 | No | ||
| 80 | ATAD2 | 2268 2256 | 8840 | 0.000 | 0.2883 | No | ||
| 81 | CXXC5 | 12523 | 8941 | 0.000 | 0.2829 | No | ||
| 82 | VLDLR | 5858 3763 912 3682 | 9237 | -0.000 | 0.2669 | No | ||
| 83 | ITGA9 | 19280 | 9390 | -0.001 | 0.2587 | No | ||
| 84 | RGMA | 18213 | 9493 | -0.001 | 0.2533 | No | ||
| 85 | RFX4 | 7721 3410 | 9495 | -0.001 | 0.2533 | No | ||
| 86 | EPAS1 | 23144 | 9649 | -0.001 | 0.2450 | No | ||
| 87 | KCNMA1 | 4947 | 9752 | -0.001 | 0.2396 | No | ||
| 88 | PLXNA1 | 5268 | 9964 | -0.002 | 0.2283 | No | ||
| 89 | SPTLC1 | 9879 3192 | 10021 | -0.002 | 0.2253 | No | ||
| 90 | WNT5A | 22066 5880 | 10268 | -0.002 | 0.2121 | No | ||
| 91 | ARHGEF17 | 17737 | 10429 | -0.002 | 0.2036 | No | ||
| 92 | DNAJA2 | 12133 18806 | 10432 | -0.002 | 0.2037 | No | ||
| 93 | MMP24 | 5110 | 10539 | -0.003 | 0.1981 | No | ||
| 94 | A2BP1 | 11212 1652 1689 | 10571 | -0.003 | 0.1966 | No | ||
| 95 | PTPLB | 22273 | 10796 | -0.003 | 0.1846 | No | ||
| 96 | NTN4 | 19903 | 10998 | -0.003 | 0.1739 | No | ||
| 97 | MINK1 | 20802 | 11160 | -0.004 | 0.1654 | No | ||
| 98 | RB1CC1 | 4486 14299 | 11284 | -0.004 | 0.1590 | No | ||
| 99 | RARB | 3011 21915 | 11666 | -0.005 | 0.1387 | No | ||
| 100 | NR2C2 | 10216 | 11854 | -0.005 | 0.1289 | No | ||
| 101 | RAP1A | 8467 | 12034 | -0.006 | 0.1195 | No | ||
| 102 | OSBPL8 | 6198 | 12124 | -0.006 | 0.1151 | No | ||
| 103 | SF4 | 18593 | 12126 | -0.006 | 0.1154 | No | ||
| 104 | DDHD1 | 497 496 4339 | 12201 | -0.006 | 0.1118 | No | ||
| 105 | ADCYAP1 | 4348 | 12612 | -0.007 | 0.0900 | No | ||
| 106 | SYNC1 | 16070 | 12972 | -0.008 | 0.0711 | No | ||
| 107 | RAB22A | 2888 5343 | 13195 | -0.009 | 0.0596 | No | ||
| 108 | TOX | 15943 | 13343 | -0.010 | 0.0523 | No | ||
| 109 | DLL4 | 7040 | 13405 | -0.010 | 0.0496 | No | ||
| 110 | TIPARP | 15576 | 13454 | -0.010 | 0.0476 | No | ||
| 111 | ZBTB11 | 11309 | 13457 | -0.010 | 0.0481 | No | ||
| 112 | PLAG1 | 7148 15949 12161 | 13525 | -0.011 | 0.0452 | No | ||
| 113 | CREB5 | 10551 | 13526 | -0.011 | 0.0458 | No | ||
| 114 | LHX6 | 9275 2914 | 13604 | -0.011 | 0.0423 | No | ||
| 115 | EMX2 | 23806 | 13605 | -0.011 | 0.0430 | No | ||
| 116 | ABCA1 | 15881 | 13652 | -0.011 | 0.0412 | No | ||
| 117 | SS18L1 | 2823 14712 | 13680 | -0.012 | 0.0404 | No | ||
| 118 | PAK7 | 14417 | 13716 | -0.012 | 0.0393 | No | ||
| 119 | SNF1LK | 23033 | 14153 | -0.015 | 0.0165 | No | ||
| 120 | TARDBP | 10520 6066 | 14555 | -0.018 | -0.0041 | No | ||
| 121 | UHMK1 | 4038 4957 | 14656 | -0.019 | -0.0084 | No | ||
| 122 | GAB2 | 1821 18184 2025 | 14705 | -0.019 | -0.0098 | No | ||
| 123 | FRAS1 | 16788 3589 | 14782 | -0.020 | -0.0127 | No | ||
| 124 | CYP26B1 | 17093 | 14783 | -0.020 | -0.0115 | No | ||
| 125 | RKHD2 | 23520 633 | 14807 | -0.020 | -0.0115 | No | ||
| 126 | FGD5 | 10555 17352 | 14861 | -0.021 | -0.0131 | No | ||
| 127 | CDH2 | 1963 8727 8726 4508 | 14992 | -0.023 | -0.0187 | No | ||
| 128 | MKRN1 | 17178 | 15239 | -0.027 | -0.0304 | No | ||
| 129 | MTUS1 | 4162 158 | 15436 | -0.030 | -0.0392 | No | ||
| 130 | PHF15 | 20467 8050 | 15517 | -0.032 | -0.0416 | No | ||
| 131 | IQWD1 | 13778 | 15580 | -0.034 | -0.0429 | No | ||
| 132 | RABGAP1 | 10439 | 15604 | -0.035 | -0.0420 | No | ||
| 133 | IRF2 | 18621 | 15738 | -0.038 | -0.0469 | No | ||
| 134 | ARF1 | 64 | 15748 | -0.038 | -0.0451 | No | ||
| 135 | ZNF385 | 22106 2297 | 15941 | -0.045 | -0.0527 | No | ||
| 136 | PBX3 | 9531 2714 2705 | 16017 | -0.048 | -0.0539 | No | ||
| 137 | ATXN7L2 | 15195 | 16386 | -0.064 | -0.0699 | No | ||
| 138 | ASF1B | 18550 | 16431 | -0.066 | -0.0683 | No | ||
| 139 | LEFTY1 | 8877 | 16470 | -0.068 | -0.0663 | No | ||
| 140 | CREBL1 | 23274 | 16489 | -0.069 | -0.0631 | No | ||
| 141 | PRKACB | 15140 | 16539 | -0.071 | -0.0614 | No | ||
| 142 | MTCH2 | 14954 7120 | 16558 | -0.072 | -0.0580 | No | ||
| 143 | AK2 | 8563 2479 16073 | 16788 | -0.089 | -0.0650 | No | ||
| 144 | RAB11A | 7013 | 16896 | -0.096 | -0.0650 | No | ||
| 145 | NT5C3 | 17136 | 16899 | -0.096 | -0.0592 | No | ||
| 146 | TUBG1 | 20662 | 17350 | -0.140 | -0.0751 | No | ||
| 147 | CDCA7 | 12437 | 17538 | -0.160 | -0.0755 | No | ||
| 148 | VAMP3 | 15660 | 17668 | -0.176 | -0.0718 | No | ||
| 149 | KPNB1 | 20274 | 17697 | -0.180 | -0.0624 | No | ||
| 150 | CDC25B | 14841 | 17982 | -0.234 | -0.0635 | No | ||
| 151 | TNFAIP1 | 20333 | 18066 | -0.263 | -0.0520 | No | ||
| 152 | DPP4 | 14579 | 18167 | -0.297 | -0.0393 | No | ||
| 153 | PAPOLA | 5261 9585 | 18279 | -0.376 | -0.0225 | No | ||
| 154 | CCND2 | 16987 | 18447 | -0.669 | 0.0092 | No |