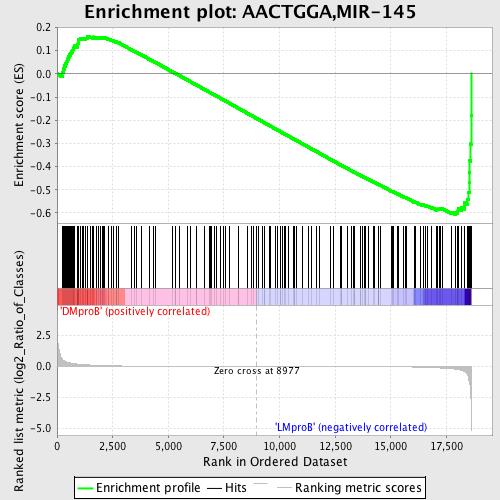
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | AACTGGA,MIR-145 |
| Enrichment Score (ES) | -0.6064394 |
| Normalized Enrichment Score (NES) | -1.416001 |
| Nominal p-value | 0.018223235 |
| FDR q-value | 0.7438662 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARHGAP24 | 16778 3655 | 233 | 0.570 | 0.0074 | No | ||
| 2 | FOXO1A | 7124 | 282 | 0.488 | 0.0220 | No | ||
| 3 | NEDD9 | 3206 21483 | 320 | 0.441 | 0.0355 | No | ||
| 4 | TM9SF4 | 14786 2830 8279 | 391 | 0.375 | 0.0450 | No | ||
| 5 | CBFB | 3769 18499 | 441 | 0.343 | 0.0544 | No | ||
| 6 | PLAGL2 | 7055 | 461 | 0.334 | 0.0651 | No | ||
| 7 | KLF5 | 4456 | 502 | 0.312 | 0.0740 | No | ||
| 8 | RASA2 | 4329 | 539 | 0.293 | 0.0823 | No | ||
| 9 | MAP3K3 | 20626 | 595 | 0.273 | 0.0890 | No | ||
| 10 | REV3L | 20050 | 639 | 0.258 | 0.0958 | No | ||
| 11 | BACH2 | 8648 | 676 | 0.247 | 0.1025 | No | ||
| 12 | CUGBP2 | 4687 8923 | 716 | 0.234 | 0.1086 | No | ||
| 13 | H2AFX | 19477 | 753 | 0.223 | 0.1145 | No | ||
| 14 | SENP2 | 7990 | 762 | 0.220 | 0.1218 | No | ||
| 15 | CHKB | 4518 | 897 | 0.193 | 0.1213 | No | ||
| 16 | SMAD3 | 19084 | 925 | 0.187 | 0.1265 | No | ||
| 17 | NFE2L1 | 9457 | 937 | 0.186 | 0.1324 | No | ||
| 18 | ADD3 | 23812 3694 | 971 | 0.179 | 0.1369 | No | ||
| 19 | CCNL1 | 15325 | 973 | 0.179 | 0.1432 | No | ||
| 20 | RNF31 | 11206 | 974 | 0.178 | 0.1495 | No | ||
| 21 | CD47 | 4933 | 1038 | 0.169 | 0.1520 | No | ||
| 22 | BAZ2A | 4371 | 1144 | 0.154 | 0.1517 | No | ||
| 23 | ZFYVE26 | 9999 | 1197 | 0.146 | 0.1540 | No | ||
| 24 | HIC2 | 7185 | 1283 | 0.135 | 0.1542 | No | ||
| 25 | PLCL2 | 5902 | 1355 | 0.125 | 0.1547 | No | ||
| 26 | FLI1 | 4729 | 1367 | 0.124 | 0.1585 | No | ||
| 27 | BLOC1S2 | 23664 | 1371 | 0.124 | 0.1627 | No | ||
| 28 | SLC25A25 | 10431 | 1520 | 0.108 | 0.1585 | No | ||
| 29 | ACSL4 | 24043 11936 24042 2614 6940 | 1611 | 0.101 | 0.1571 | No | ||
| 30 | USP52 | 3397 19841 | 1642 | 0.098 | 0.1590 | No | ||
| 31 | AP1G1 | 4392 | 1753 | 0.089 | 0.1561 | No | ||
| 32 | XRN1 | 10797 | 1774 | 0.088 | 0.1581 | No | ||
| 33 | RAB14 | 12685 | 1880 | 0.081 | 0.1553 | No | ||
| 34 | EIF4EBP2 | 4662 | 1929 | 0.077 | 0.1554 | No | ||
| 35 | MRPL48 | 3328 17739 2035 2242 3883 | 1937 | 0.077 | 0.1577 | No | ||
| 36 | ZDHHC9 | 24163 | 2057 | 0.068 | 0.1537 | No | ||
| 37 | MAP4K4 | 11241 | 2059 | 0.068 | 0.1560 | No | ||
| 38 | LHFPL2 | 21584 | 2101 | 0.065 | 0.1561 | No | ||
| 39 | ZFYVE9 | 10501 2449 | 2151 | 0.063 | 0.1556 | No | ||
| 40 | SRGAP2 | 13836 17045 6433 17046 | 2300 | 0.056 | 0.1496 | No | ||
| 41 | ZBTB10 | 10468 | 2323 | 0.055 | 0.1503 | No | ||
| 42 | SCAMP3 | 15543 | 2457 | 0.049 | 0.1448 | No | ||
| 43 | DYRK1A | 4649 | 2521 | 0.046 | 0.1431 | No | ||
| 44 | ABR | 8468 20346 1476 1429 | 2651 | 0.041 | 0.1375 | No | ||
| 45 | RASA1 | 10174 | 2654 | 0.041 | 0.1389 | No | ||
| 46 | ADPGK | 13003 7788 19422 | 2766 | 0.037 | 0.1342 | No | ||
| 47 | LOX | 23438 | 3360 | 0.024 | 0.1029 | No | ||
| 48 | ADAM19 | 8548 | 3483 | 0.023 | 0.0971 | No | ||
| 49 | TIRAP | 3075 19187 | 3576 | 0.021 | 0.0929 | No | ||
| 50 | PTP4A2 | 9659 2463 2518 5324 | 3787 | 0.018 | 0.0821 | No | ||
| 51 | ARPC5 | 12580 7487 | 4151 | 0.015 | 0.0630 | No | ||
| 52 | ACVR2A | 8542 | 4335 | 0.013 | 0.0535 | No | ||
| 53 | CITED2 | 5118 14477 | 4423 | 0.013 | 0.0493 | No | ||
| 54 | SCN3A | 14573 | 5187 | 0.009 | 0.0082 | No | ||
| 55 | TNFRSF11B | 22296 | 5313 | 0.009 | 0.0018 | No | ||
| 56 | PPP3CA | 1863 5284 | 5331 | 0.008 | 0.0012 | No | ||
| 57 | FKBP3 | 21056 | 5483 | 0.008 | -0.0068 | No | ||
| 58 | SLITRK4 | 24144 | 5870 | 0.007 | -0.0274 | No | ||
| 59 | SLC4A4 | 16807 12035 7036 | 6011 | 0.006 | -0.0348 | No | ||
| 60 | RIN2 | 14817 | 6256 | 0.005 | -0.0478 | No | ||
| 61 | BCR | 8478 3384 19990 | 6282 | 0.005 | -0.0490 | No | ||
| 62 | CFL2 | 2155 21069 | 6622 | 0.005 | -0.0672 | No | ||
| 63 | ACTG1 | 8535 | 6851 | 0.004 | -0.0794 | No | ||
| 64 | CABP1 | 16412 | 6896 | 0.004 | -0.0817 | No | ||
| 65 | TBPL1 | 19805 | 6940 | 0.004 | -0.0839 | No | ||
| 66 | SLC6A15 | 8335 3321 | 7088 | 0.004 | -0.0917 | No | ||
| 67 | GABARAPL2 | 8211 13552 | 7178 | 0.003 | -0.0964 | No | ||
| 68 | AFF2 | 8977 | 7322 | 0.003 | -0.1040 | No | ||
| 69 | JPH1 | 13994 | 7493 | 0.003 | -0.1132 | No | ||
| 70 | LENG8 | 6114 | 7575 | 0.003 | -0.1175 | No | ||
| 71 | BAAT | 15885 | 7750 | 0.002 | -0.1268 | No | ||
| 72 | FNDC3A | 6672 21756 | 8149 | 0.001 | -0.1483 | No | ||
| 73 | CLCA3 | 6224 | 8152 | 0.001 | -0.1484 | No | ||
| 74 | MYO5A | 19378 2978 | 8559 | 0.001 | -0.1704 | No | ||
| 75 | SRGAP1 | 19615 | 8733 | 0.000 | -0.1797 | No | ||
| 76 | CAMK1D | 5977 2948 | 8821 | 0.000 | -0.1844 | No | ||
| 77 | ACTR3 | 13166 | 8962 | 0.000 | -0.1920 | No | ||
| 78 | SEMA3A | 16919 | 8963 | 0.000 | -0.1920 | No | ||
| 79 | YES1 | 5930 | 9040 | -0.000 | -0.1961 | No | ||
| 80 | GGTL3 | 14381 | 9246 | -0.000 | -0.2072 | No | ||
| 81 | SOX11 | 5477 | 9302 | -0.001 | -0.2102 | No | ||
| 82 | SLITRK6 | 21724 | 9305 | -0.001 | -0.2102 | No | ||
| 83 | IRS1 | 4925 | 9553 | -0.001 | -0.2236 | No | ||
| 84 | TRIM2 | 13485 8157 | 9587 | -0.001 | -0.2254 | No | ||
| 85 | UXS1 | 13965 | 9809 | -0.001 | -0.2373 | No | ||
| 86 | KIAA1715 | 7629 | 9889 | -0.001 | -0.2415 | No | ||
| 87 | FBXO28 | 7508 12612 | 10020 | -0.002 | -0.2485 | No | ||
| 88 | SPATS2 | 13044 | 10045 | -0.002 | -0.2497 | No | ||
| 89 | KIF21A | 4952 | 10137 | -0.002 | -0.2546 | No | ||
| 90 | EYA3 | 8936 2352 | 10205 | -0.002 | -0.2582 | No | ||
| 91 | CSMD3 | 10735 7938 | 10271 | -0.002 | -0.2616 | No | ||
| 92 | SEMA6A | 23439 9802 | 10413 | -0.002 | -0.2692 | No | ||
| 93 | PXN | 5339 3573 | 10622 | -0.003 | -0.2803 | No | ||
| 94 | PTGFR | 15138 | 10661 | -0.003 | -0.2823 | No | ||
| 95 | PLCE1 | 13156 | 10690 | -0.003 | -0.2837 | No | ||
| 96 | ATXN2 | 9780 3506 | 10767 | -0.003 | -0.2877 | No | ||
| 97 | SLC1A2 | 5453 | 11022 | -0.004 | -0.3014 | No | ||
| 98 | DACH1 | 4595 3371 | 11296 | -0.004 | -0.3160 | No | ||
| 99 | ZFP2 | 10385 1366 1474 | 11449 | -0.004 | -0.3241 | No | ||
| 100 | AMOTL2 | 19339 | 11664 | -0.005 | -0.3355 | No | ||
| 101 | SACM1L | 19253 | 11790 | -0.005 | -0.3421 | No | ||
| 102 | PHLDB2 | 22581 | 12272 | -0.006 | -0.3679 | No | ||
| 103 | MPP5 | 21230 7078 | 12417 | -0.007 | -0.3755 | No | ||
| 104 | QKI | 23128 5340 | 12720 | -0.008 | -0.3916 | No | ||
| 105 | RAPH1 | 19110 | 12772 | -0.008 | -0.3941 | No | ||
| 106 | ANGPT2 | 14885 3783 18923 | 12795 | -0.008 | -0.3950 | No | ||
| 107 | PAPD4 | 8299 8300 4146 | 13048 | -0.009 | -0.4083 | No | ||
| 108 | SBF2 | 11485 | 13212 | -0.009 | -0.4168 | No | ||
| 109 | RTKN | 17394 1156 | 13317 | -0.010 | -0.4221 | No | ||
| 110 | ERG | 1686 8915 | 13370 | -0.010 | -0.4246 | No | ||
| 111 | ABLIM2 | 16866 3605 | 13376 | -0.010 | -0.4245 | No | ||
| 112 | INHBB | 13858 | 13618 | -0.011 | -0.4372 | No | ||
| 113 | ABCA1 | 15881 | 13652 | -0.011 | -0.4386 | No | ||
| 114 | PAK7 | 14417 | 13716 | -0.012 | -0.4416 | No | ||
| 115 | FZD7 | 14242 | 13801 | -0.012 | -0.4457 | No | ||
| 116 | EFNA3 | 15278 | 13880 | -0.013 | -0.4494 | No | ||
| 117 | BSN | 18997 | 13882 | -0.013 | -0.4490 | No | ||
| 118 | LRRC16 | 21515 21514 | 13978 | -0.014 | -0.4537 | No | ||
| 119 | KIF1B | 15671 2476 4950 15673 | 14222 | -0.015 | -0.4663 | No | ||
| 120 | RGS7 | 13743 | 14270 | -0.015 | -0.4683 | No | ||
| 121 | CSTF3 | 10449 6002 | 14443 | -0.017 | -0.4771 | No | ||
| 122 | RSPO1 | 16087 | 14462 | -0.017 | -0.4774 | No | ||
| 123 | FXN | 23709 | 14465 | -0.017 | -0.4770 | No | ||
| 124 | SOX9 | 20611 | 14535 | -0.018 | -0.4801 | No | ||
| 125 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 15048 | -0.024 | -0.5070 | No | ||
| 126 | DLX6 | 17527 | 15072 | -0.024 | -0.5074 | No | ||
| 127 | TGFBR2 | 2994 3001 10167 | 15132 | -0.025 | -0.5097 | No | ||
| 128 | AKAP12 | 17600 | 15298 | -0.028 | -0.5177 | No | ||
| 129 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 15324 | -0.028 | -0.5180 | No | ||
| 130 | GLIS1 | 16150 | 15566 | -0.034 | -0.5299 | No | ||
| 131 | YTHDF2 | 5623 5624 | 15667 | -0.036 | -0.5341 | No | ||
| 132 | NUDT4 | 19639 | 15683 | -0.036 | -0.5336 | No | ||
| 133 | RBPMS | 5369 | 16056 | -0.049 | -0.5520 | No | ||
| 134 | ACVR1B | 4335 22353 | 16103 | -0.051 | -0.5527 | No | ||
| 135 | DNAJB11 | 22813 | 16321 | -0.061 | -0.5623 | No | ||
| 136 | DAG1 | 18996 8837 | 16459 | -0.067 | -0.5674 | No | ||
| 137 | SSH2 | 6202 | 16461 | -0.067 | -0.5651 | No | ||
| 138 | NET1 | 7100 | 16555 | -0.072 | -0.5676 | No | ||
| 139 | CDK6 | 16600 | 16668 | -0.079 | -0.5709 | No | ||
| 140 | SPOP | 5498 | 16848 | -0.093 | -0.5773 | No | ||
| 141 | FLNB | 13742 | 17059 | -0.109 | -0.5849 | No | ||
| 142 | NAP1L1 | 19882 3427 330 9485 5193 | 17092 | -0.111 | -0.5827 | No | ||
| 143 | EIF4B | 13279 7979 | 17172 | -0.119 | -0.5828 | No | ||
| 144 | SKP1A | 20889 | 17233 | -0.126 | -0.5816 | No | ||
| 145 | RASSF5 | 13837 | 17324 | -0.136 | -0.5817 | No | ||
| 146 | GPHN | 6492 21232 | 17734 | -0.184 | -0.5973 | No | ||
| 147 | NEDD4L | 8192 | 17903 | -0.215 | -0.5989 | Yes | ||
| 148 | AP3S1 | 8604 | 17998 | -0.239 | -0.5956 | Yes | ||
| 149 | ACBD3 | 14025 | 18018 | -0.245 | -0.5880 | Yes | ||
| 150 | TAGLN2 | 14043 | 18042 | -0.255 | -0.5803 | Yes | ||
| 151 | IVNS1ABP | 4384 4050 | 18190 | -0.312 | -0.5772 | Yes | ||
| 152 | ACACA | 309 4209 | 18298 | -0.389 | -0.5693 | Yes | ||
| 153 | EVA1 | 19472 | 18324 | -0.411 | -0.5562 | Yes | ||
| 154 | SPSB4 | 19035 | 18436 | -0.613 | -0.5406 | Yes | ||
| 155 | GPIAP1 | 2819 7014 | 18498 | -0.964 | -0.5100 | Yes | ||
| 156 | DUSP6 | 19891 3399 | 18525 | -1.221 | -0.4684 | Yes | ||
| 157 | PDCD4 | 5232 23816 | 18530 | -1.281 | -0.4235 | Yes | ||
| 158 | EIF4A2 | 4660 1679 1645 | 18549 | -1.405 | -0.3750 | Yes | ||
| 159 | AKAP9 | 16604 16603 4150 8303 | 18594 | -2.133 | -0.3023 | Yes | ||
| 160 | ACTB | 8534 337 337 338 | 18610 | -3.484 | -0.1804 | Yes | ||
| 161 | DPYSL2 | 3122 4561 8787 | 18616 | -5.131 | -0.0000 | Yes |

