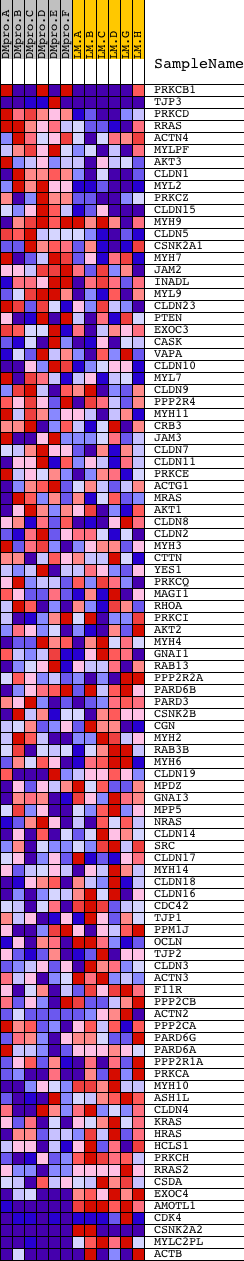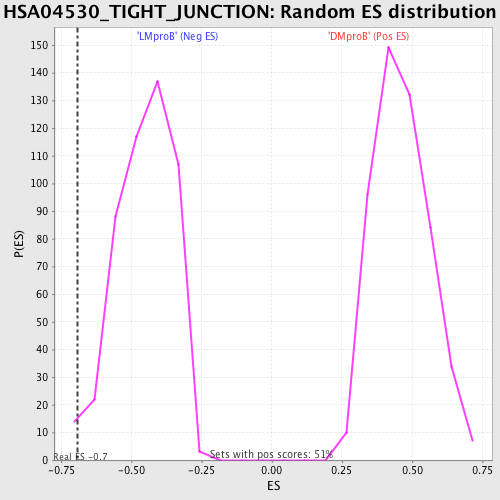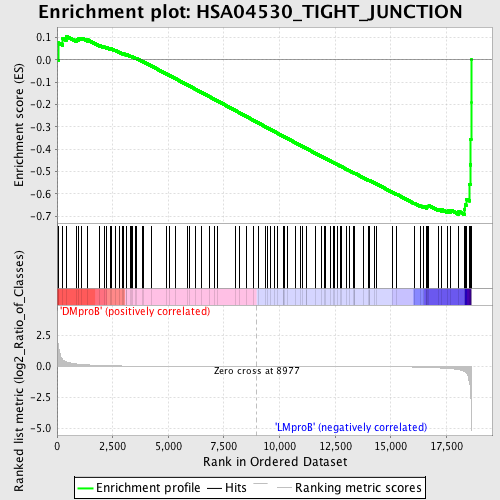
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04530_TIGHT_JUNCTION |
| Enrichment Score (ES) | -0.69191337 |
| Normalized Enrichment Score (NES) | -1.5307693 |
| Nominal p-value | 0.014344262 |
| FDR q-value | 0.61373067 |
| FWER p-Value | 0.988 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRKCB1 | 1693 9574 | 62 | 1.449 | 0.0757 | No | ||
| 2 | TJP3 | 19677 3298 | 255 | 0.528 | 0.0941 | No | ||
| 3 | PRKCD | 21897 | 406 | 0.366 | 0.1060 | No | ||
| 4 | RRAS | 18256 | 891 | 0.194 | 0.0904 | No | ||
| 5 | ACTN4 | 17905 1798 1983 | 975 | 0.178 | 0.0957 | No | ||
| 6 | MYLPF | 18069 | 1105 | 0.158 | 0.0973 | No | ||
| 7 | AKT3 | 13739 982 | 1372 | 0.123 | 0.0897 | No | ||
| 8 | CLDN1 | 22623 | 1910 | 0.078 | 0.0649 | No | ||
| 9 | MYL2 | 16713 | 2114 | 0.064 | 0.0575 | No | ||
| 10 | PRKCZ | 5260 | 2237 | 0.059 | 0.0541 | No | ||
| 11 | CLDN15 | 16667 3572 | 2403 | 0.051 | 0.0480 | No | ||
| 12 | MYH9 | 2252 2244 | 2429 | 0.050 | 0.0494 | No | ||
| 13 | CLDN5 | 22826 | 2606 | 0.043 | 0.0422 | No | ||
| 14 | CSNK2A1 | 14797 | 2823 | 0.036 | 0.0325 | No | ||
| 15 | MYH7 | 21828 4709 | 2950 | 0.033 | 0.0275 | No | ||
| 16 | JAM2 | 7428 22718 12519 | 2989 | 0.032 | 0.0272 | No | ||
| 17 | INADL | 2517 2403 165 2347 166 | 3102 | 0.029 | 0.0227 | No | ||
| 18 | MYL9 | 14767 2659 | 3124 | 0.029 | 0.0232 | No | ||
| 19 | CLDN23 | 18893 | 3279 | 0.026 | 0.0163 | No | ||
| 20 | PTEN | 5305 | 3348 | 0.025 | 0.0140 | No | ||
| 21 | EXOC3 | 9986 | 3374 | 0.024 | 0.0139 | No | ||
| 22 | CASK | 24184 2618 2604 | 3528 | 0.022 | 0.0069 | No | ||
| 23 | VAPA | 22908 | 3571 | 0.021 | 0.0057 | No | ||
| 24 | CLDN10 | 21935 2785 2601 2720 7188 | 3853 | 0.018 | -0.0085 | No | ||
| 25 | MYL7 | 9437 | 3896 | 0.017 | -0.0098 | No | ||
| 26 | CLDN9 | 24470 | 4260 | 0.014 | -0.0286 | No | ||
| 27 | PPP2R4 | 15050 2910 | 4925 | 0.010 | -0.0639 | No | ||
| 28 | MYH11 | 22656 | 5062 | 0.010 | -0.0708 | No | ||
| 29 | CRB3 | 23176 | 5318 | 0.008 | -0.0841 | No | ||
| 30 | JAM3 | 8197 | 5846 | 0.007 | -0.1122 | No | ||
| 31 | CLDN7 | 20817 | 5950 | 0.006 | -0.1174 | No | ||
| 32 | CLDN11 | 15616 | 6228 | 0.006 | -0.1320 | No | ||
| 33 | PRKCE | 9575 | 6472 | 0.005 | -0.1449 | No | ||
| 34 | ACTG1 | 8535 | 6851 | 0.004 | -0.1651 | No | ||
| 35 | MRAS | 9409 | 7054 | 0.004 | -0.1758 | No | ||
| 36 | AKT1 | 8568 | 7187 | 0.003 | -0.1827 | No | ||
| 37 | CLDN8 | 22549 1747 | 8015 | 0.002 | -0.2273 | No | ||
| 38 | CLDN2 | 4527 | 8178 | 0.001 | -0.2360 | No | ||
| 39 | MYH3 | 20839 | 8513 | 0.001 | -0.2540 | No | ||
| 40 | CTTN | 8817 4575 1035 8818 | 8823 | 0.000 | -0.2706 | No | ||
| 41 | YES1 | 5930 | 9040 | -0.000 | -0.2823 | No | ||
| 42 | PRKCQ | 2873 2831 | 9370 | -0.001 | -0.3000 | No | ||
| 43 | MAGI1 | 19828 4816 1179 | 9454 | -0.001 | -0.3045 | No | ||
| 44 | RHOA | 8624 4409 4410 | 9583 | -0.001 | -0.3113 | No | ||
| 45 | PRKCI | 9576 | 9792 | -0.001 | -0.3225 | No | ||
| 46 | AKT2 | 4365 4366 | 9914 | -0.001 | -0.3289 | No | ||
| 47 | MYH4 | 20837 | 10175 | -0.002 | -0.3429 | No | ||
| 48 | GNAI1 | 9024 | 10178 | -0.002 | -0.3429 | No | ||
| 49 | RAB13 | 15535 | 10207 | -0.002 | -0.3443 | No | ||
| 50 | PPP2R2A | 3222 21774 | 10346 | -0.002 | -0.3516 | No | ||
| 51 | PARD6B | 12209 | 10701 | -0.003 | -0.3705 | No | ||
| 52 | PARD3 | 8212 13553 | 10938 | -0.003 | -0.3831 | No | ||
| 53 | CSNK2B | 23008 | 11024 | -0.004 | -0.3875 | No | ||
| 54 | CGN | 12896 12897 7701 | 11194 | -0.004 | -0.3964 | No | ||
| 55 | MYH2 | 20838 | 11220 | -0.004 | -0.3975 | No | ||
| 56 | RAB3B | 16141 | 11623 | -0.005 | -0.4190 | No | ||
| 57 | MYH6 | 9436 | 11865 | -0.005 | -0.4317 | No | ||
| 58 | CLDN19 | 16105 472 | 12016 | -0.006 | -0.4395 | No | ||
| 59 | MPDZ | 2497 2366 9406 | 12074 | -0.006 | -0.4423 | No | ||
| 60 | GNAI3 | 15198 | 12266 | -0.006 | -0.4522 | No | ||
| 61 | MPP5 | 21230 7078 | 12417 | -0.007 | -0.4600 | No | ||
| 62 | NRAS | 5191 | 12478 | -0.007 | -0.4628 | No | ||
| 63 | CLDN14 | 12081 | 12585 | -0.007 | -0.4681 | No | ||
| 64 | SRC | 5507 | 12717 | -0.008 | -0.4748 | No | ||
| 65 | CLDN17 | 10757 | 12801 | -0.008 | -0.4789 | No | ||
| 66 | MYH14 | 17845 | 12989 | -0.008 | -0.4885 | No | ||
| 67 | CLDN18 | 19028 | 13147 | -0.009 | -0.4965 | No | ||
| 68 | CLDN16 | 22802 | 13314 | -0.010 | -0.5049 | No | ||
| 69 | CDC42 | 4503 8722 4504 2465 | 13354 | -0.010 | -0.5065 | No | ||
| 70 | TJP1 | 17803 | 13375 | -0.010 | -0.5070 | No | ||
| 71 | PPM1J | 15466 12971 | 13751 | -0.012 | -0.5266 | No | ||
| 72 | OCLN | 21368 | 14002 | -0.014 | -0.5393 | No | ||
| 73 | TJP2 | 5761 3699 | 14030 | -0.014 | -0.5400 | No | ||
| 74 | CLDN3 | 16680 | 14033 | -0.014 | -0.5394 | No | ||
| 75 | ACTN3 | 23967 8540 | 14246 | -0.015 | -0.5500 | No | ||
| 76 | F11R | 9199 | 14365 | -0.016 | -0.5555 | No | ||
| 77 | PPP2CB | 18636 | 15065 | -0.024 | -0.5920 | No | ||
| 78 | ACTN2 | 21545 | 15249 | -0.027 | -0.6004 | No | ||
| 79 | PPP2CA | 20890 | 16076 | -0.050 | -0.6423 | No | ||
| 80 | PARD6G | 23503 | 16354 | -0.062 | -0.6538 | No | ||
| 81 | PARD6A | 12142 | 16488 | -0.068 | -0.6573 | No | ||
| 82 | PPP2R1A | 11951 | 16611 | -0.076 | -0.6597 | No | ||
| 83 | PRKCA | 20174 | 16623 | -0.077 | -0.6561 | No | ||
| 84 | MYH10 | 8077 | 16669 | -0.079 | -0.6542 | No | ||
| 85 | ASH1L | 5311 15546 15547 | 16714 | -0.083 | -0.6521 | No | ||
| 86 | CLDN4 | 8747 | 17153 | -0.118 | -0.6693 | No | ||
| 87 | KRAS | 9247 | 17295 | -0.133 | -0.6697 | No | ||
| 88 | HRAS | 4868 | 17569 | -0.163 | -0.6755 | No | ||
| 89 | HCLS1 | 22770 | 17688 | -0.178 | -0.6721 | No | ||
| 90 | PRKCH | 21246 | 18055 | -0.260 | -0.6777 | Yes | ||
| 91 | RRAS2 | 12431 | 18302 | -0.392 | -0.6696 | Yes | ||
| 92 | CSDA | 1163 16966 1134 | 18334 | -0.427 | -0.6480 | Yes | ||
| 93 | EXOC4 | 17493 | 18389 | -0.498 | -0.6237 | Yes | ||
| 94 | AMOTL1 | 19242 | 18540 | -1.348 | -0.5582 | Yes | ||
| 95 | CDK4 | 3424 19859 | 18570 | -1.651 | -0.4697 | Yes | ||
| 96 | CSNK2A2 | 4567 8808 | 18591 | -2.083 | -0.3572 | Yes | ||
| 97 | MYLC2PL | 16670 | 18607 | -3.084 | -0.1897 | Yes | ||
| 98 | ACTB | 8534 337 337 338 | 18610 | -3.484 | 0.0003 | Yes |