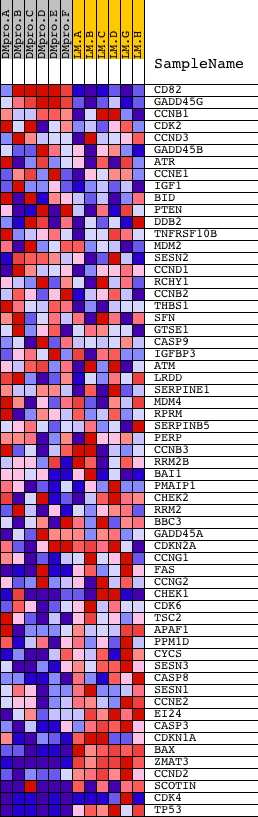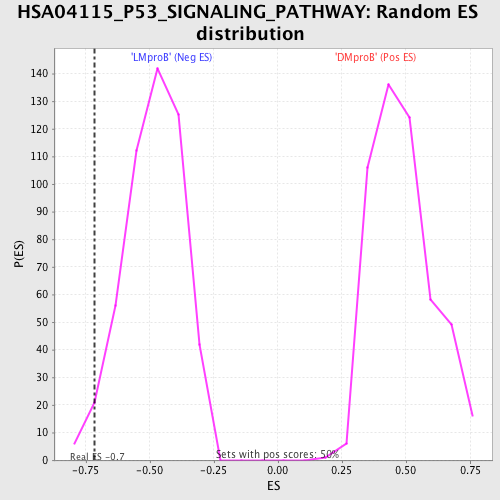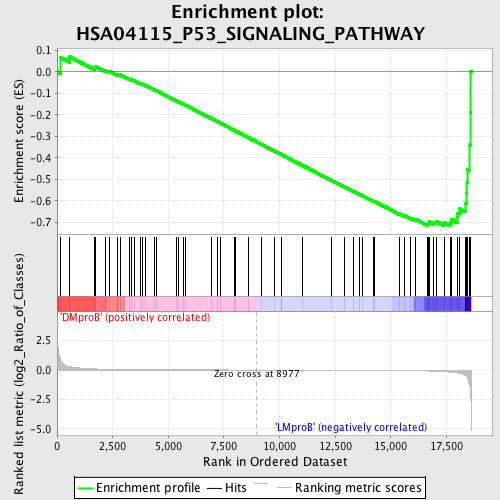
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA04115_P53_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.7170648 |
| Normalized Enrichment Score (NES) | -1.4826626 |
| Nominal p-value | 0.025793651 |
| FDR q-value | 0.8773825 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CD82 | 8720 | 163 | 0.809 | 0.0656 | No | ||
| 2 | GADD45G | 21637 | 573 | 0.279 | 0.0692 | No | ||
| 3 | CCNB1 | 11201 21362 | 1697 | 0.093 | 0.0173 | No | ||
| 4 | CDK2 | 3438 3373 19592 3322 | 1735 | 0.090 | 0.0236 | No | ||
| 5 | CCND3 | 4489 4490 | 2174 | 0.062 | 0.0056 | No | ||
| 6 | GADD45B | 19935 | 2352 | 0.053 | 0.0010 | No | ||
| 7 | ATR | 19349 | 2705 | 0.040 | -0.0143 | No | ||
| 8 | CCNE1 | 17857 | 2715 | 0.039 | -0.0112 | No | ||
| 9 | IGF1 | 3352 9156 3409 | 2839 | 0.035 | -0.0146 | No | ||
| 10 | BID | 17017 435 | 3254 | 0.026 | -0.0345 | No | ||
| 11 | PTEN | 5305 | 3348 | 0.025 | -0.0372 | No | ||
| 12 | DDB2 | 2721 2878 14525 2661 | 3464 | 0.023 | -0.0413 | No | ||
| 13 | TNFRSF10B | 21971 10205 5781 10204 | 3733 | 0.019 | -0.0540 | No | ||
| 14 | MDM2 | 19620 3327 | 3839 | 0.018 | -0.0580 | No | ||
| 15 | SESN2 | 15733 | 3951 | 0.017 | -0.0625 | No | ||
| 16 | CCND1 | 4487 4488 8707 17535 | 4387 | 0.013 | -0.0847 | No | ||
| 17 | RCHY1 | 12646 | 4449 | 0.013 | -0.0868 | No | ||
| 18 | CCNB2 | 19067 | 5380 | 0.008 | -0.1362 | No | ||
| 19 | THBS1 | 5748 14910 | 5472 | 0.008 | -0.1404 | No | ||
| 20 | SFN | 7060 12062 | 5663 | 0.007 | -0.1499 | No | ||
| 21 | GTSE1 | 11385 2170 | 5748 | 0.007 | -0.1538 | No | ||
| 22 | CASP9 | 16001 2410 2458 | 6943 | 0.004 | -0.2178 | No | ||
| 23 | IGFBP3 | 4898 | 7202 | 0.003 | -0.2314 | No | ||
| 24 | ATM | 2976 19115 | 7358 | 0.003 | -0.2395 | No | ||
| 25 | LRDD | 17555 | 7959 | 0.002 | -0.2717 | No | ||
| 26 | SERPINE1 | 16333 | 8037 | 0.002 | -0.2757 | No | ||
| 27 | MDM4 | 5086 5085 | 8620 | 0.001 | -0.3070 | No | ||
| 28 | RPRM | 14588 | 9191 | -0.000 | -0.3377 | No | ||
| 29 | SERPINB5 | 9861 | 9780 | -0.001 | -0.3692 | No | ||
| 30 | PERP | 2259 22121 20085 | 10065 | -0.002 | -0.3844 | No | ||
| 31 | CCNB3 | 24205 | 10090 | -0.002 | -0.3855 | No | ||
| 32 | RRM2B | 6902 6903 11801 | 11023 | -0.004 | -0.4354 | No | ||
| 33 | BAI1 | 22459 | 12324 | -0.007 | -0.5049 | No | ||
| 34 | PMAIP1 | 23530 12216 7196 | 12920 | -0.008 | -0.5362 | No | ||
| 35 | CHEK2 | 16751 3587 | 13338 | -0.010 | -0.5577 | No | ||
| 36 | RRM2 | 5401 5400 | 13596 | -0.011 | -0.5705 | No | ||
| 37 | BBC3 | 18382 2044 | 13721 | -0.012 | -0.5761 | No | ||
| 38 | GADD45A | 17129 | 14234 | -0.015 | -0.6023 | No | ||
| 39 | CDKN2A | 2491 15841 | 14281 | -0.016 | -0.6034 | No | ||
| 40 | CCNG1 | 4492 1236 1390 | 15383 | -0.029 | -0.6600 | No | ||
| 41 | FAS | 23881 3750 | 15634 | -0.035 | -0.6703 | No | ||
| 42 | CCNG2 | 16790 | 15905 | -0.043 | -0.6808 | No | ||
| 43 | CHEK1 | 19181 3085 | 16130 | -0.052 | -0.6881 | No | ||
| 44 | CDK6 | 16600 | 16668 | -0.079 | -0.7098 | Yes | ||
| 45 | TSC2 | 23090 | 16672 | -0.079 | -0.7027 | Yes | ||
| 46 | APAF1 | 8606 | 16737 | -0.085 | -0.6983 | Yes | ||
| 47 | PPM1D | 20721 | 16933 | -0.098 | -0.6998 | Yes | ||
| 48 | CYCS | 8821 | 17067 | -0.110 | -0.6969 | Yes | ||
| 49 | SESN3 | 19561 | 17396 | -0.144 | -0.7014 | Yes | ||
| 50 | CASP8 | 8694 | 17662 | -0.175 | -0.6995 | Yes | ||
| 51 | SESN1 | 20045 | 17738 | -0.185 | -0.6865 | Yes | ||
| 52 | CCNE2 | 4491 | 17985 | -0.236 | -0.6781 | Yes | ||
| 53 | EI24 | 19179 | 18006 | -0.241 | -0.6571 | Yes | ||
| 54 | CASP3 | 8693 | 18077 | -0.266 | -0.6364 | Yes | ||
| 55 | CDKN1A | 4511 8729 | 18339 | -0.430 | -0.6109 | Yes | ||
| 56 | BAX | 17832 | 18406 | -0.524 | -0.5663 | Yes | ||
| 57 | ZMAT3 | 10305 | 18423 | -0.576 | -0.5141 | Yes | ||
| 58 | CCND2 | 16987 | 18447 | -0.669 | -0.4538 | Yes | ||
| 59 | SCOTIN | 3136 19299 3102 | 18533 | -1.296 | -0.3393 | Yes | ||
| 60 | CDK4 | 3424 19859 | 18570 | -1.651 | -0.1894 | Yes | ||
| 61 | TP53 | 20822 | 18592 | -2.086 | 0.0013 | Yes |