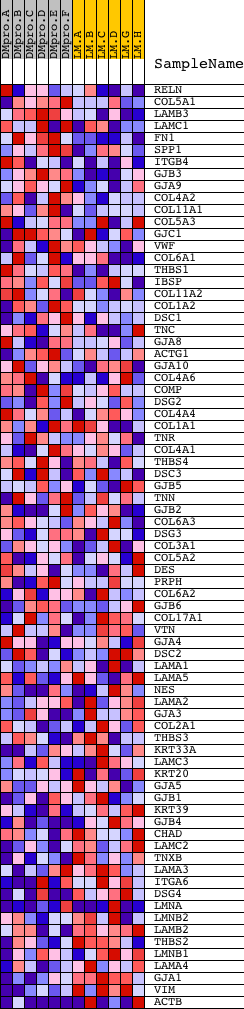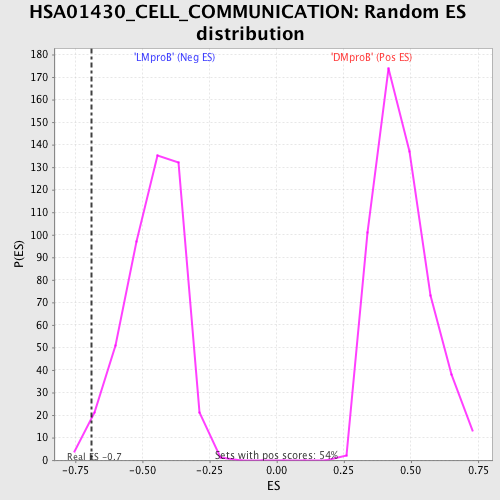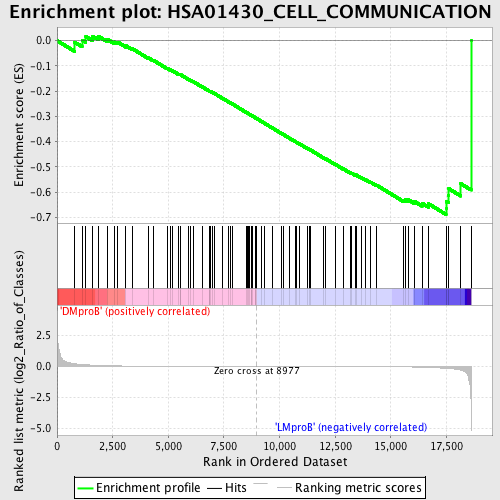
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMproB_versus_LMproB.phenotype_DMproB_versus_LMproB.cls #DMproB_versus_LMproB |
| Phenotype | phenotype_DMproB_versus_LMproB.cls#DMproB_versus_LMproB |
| Upregulated in class | LMproB |
| GeneSet | HSA01430_CELL_COMMUNICATION |
| Enrichment Score (ES) | -0.6886176 |
| Normalized Enrichment Score (NES) | -1.5011444 |
| Nominal p-value | 0.015151516 |
| FDR q-value | 0.7978441 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RELN | 5372 | 784 | 0.215 | -0.0058 | No | ||
| 2 | COL5A1 | 4544 | 1122 | 0.156 | 0.0025 | No | ||
| 3 | LAMB3 | 14007 | 1292 | 0.133 | 0.0161 | No | ||
| 4 | LAMC1 | 13805 | 1582 | 0.103 | 0.0180 | No | ||
| 5 | FN1 | 4091 4094 971 13929 | 1862 | 0.082 | 0.0169 | No | ||
| 6 | SPP1 | 5501 | 2265 | 0.058 | 0.0050 | No | ||
| 7 | ITGB4 | 9669 159 | 2592 | 0.044 | -0.0052 | No | ||
| 8 | GJB3 | 15754 | 2722 | 0.039 | -0.0055 | No | ||
| 9 | GJA9 | 14483 4781 | 3092 | 0.030 | -0.0204 | No | ||
| 10 | COL4A2 | 18688 | 3382 | 0.024 | -0.0319 | No | ||
| 11 | COL11A1 | 15442 1937 4540 8762 | 4105 | 0.015 | -0.0682 | No | ||
| 12 | COL5A3 | 19219 | 4329 | 0.013 | -0.0780 | No | ||
| 13 | GJC1 | 20256 | 4942 | 0.010 | -0.1093 | No | ||
| 14 | VWF | 17277 | 5089 | 0.009 | -0.1156 | No | ||
| 15 | COL6A1 | 19722 19721 8768 | 5206 | 0.009 | -0.1203 | No | ||
| 16 | THBS1 | 5748 14910 | 5472 | 0.008 | -0.1333 | No | ||
| 17 | IBSP | 16775 | 5523 | 0.008 | -0.1346 | No | ||
| 18 | COL11A2 | 23286 | 5536 | 0.008 | -0.1340 | No | ||
| 19 | COL1A2 | 8772 | 5903 | 0.007 | -0.1526 | No | ||
| 20 | DSC1 | 8864 | 5988 | 0.006 | -0.1561 | No | ||
| 21 | TNC | 2319 2544 2421 15861 2448 2436 | 6120 | 0.006 | -0.1622 | No | ||
| 22 | GJA8 | 15234 | 6527 | 0.005 | -0.1833 | No | ||
| 23 | ACTG1 | 8535 | 6851 | 0.004 | -0.2000 | No | ||
| 24 | GJA10 | 16077 | 6859 | 0.004 | -0.1997 | No | ||
| 25 | COL4A6 | 8248 2552 | 6905 | 0.004 | -0.2015 | No | ||
| 26 | COMP | 18589 | 6991 | 0.004 | -0.2054 | No | ||
| 27 | DSG2 | 2045 4643 | 7060 | 0.004 | -0.2085 | No | ||
| 28 | COL4A4 | 8767 | 7442 | 0.003 | -0.2285 | No | ||
| 29 | COL1A1 | 8771 | 7716 | 0.002 | -0.2429 | No | ||
| 30 | TNR | 5787 | 7788 | 0.002 | -0.2463 | No | ||
| 31 | COL4A1 | 18936 | 7897 | 0.002 | -0.2518 | No | ||
| 32 | THBS4 | 21393 | 8494 | 0.001 | -0.2838 | No | ||
| 33 | DSC3 | 23487 | 8567 | 0.001 | -0.2876 | No | ||
| 34 | GJB5 | 15753 | 8594 | 0.001 | -0.2889 | No | ||
| 35 | TNN | 11625 | 8662 | 0.001 | -0.2924 | No | ||
| 36 | GJB2 | 9018 21806 | 8735 | 0.000 | -0.2962 | No | ||
| 37 | COL6A3 | 4546 13885 13884 | 8754 | 0.000 | -0.2972 | No | ||
| 38 | DSG3 | 8866 | 8775 | 0.000 | -0.2982 | No | ||
| 39 | COL3A1 | 14253 8765 | 8905 | 0.000 | -0.3051 | No | ||
| 40 | COL5A2 | 4545 | 8923 | 0.000 | -0.3060 | No | ||
| 41 | DES | 14215 | 8940 | 0.000 | -0.3069 | No | ||
| 42 | PRPH | 5294 | 9184 | -0.000 | -0.3199 | No | ||
| 43 | COL6A2 | 19723 | 9336 | -0.001 | -0.3279 | No | ||
| 44 | GJB6 | 21805 | 9674 | -0.001 | -0.3459 | No | ||
| 45 | COL17A1 | 23647 3671 | 10079 | -0.002 | -0.3674 | No | ||
| 46 | VTN | 20756 | 10167 | -0.002 | -0.3718 | No | ||
| 47 | GJA4 | 15755 | 10464 | -0.002 | -0.3873 | No | ||
| 48 | DSC2 | 8865 | 10697 | -0.003 | -0.3993 | No | ||
| 49 | LAMA1 | 23168 | 10779 | -0.003 | -0.4032 | No | ||
| 50 | LAMA5 | 9263 | 10877 | -0.003 | -0.4078 | No | ||
| 51 | NES | 5162 9454 | 11273 | -0.004 | -0.4285 | No | ||
| 52 | LAMA2 | 19800 | 11325 | -0.004 | -0.4305 | No | ||
| 53 | GJA3 | 21807 | 11375 | -0.004 | -0.4324 | No | ||
| 54 | COL2A1 | 22145 4542 | 11992 | -0.006 | -0.4647 | No | ||
| 55 | THBS3 | 15542 1796 | 12082 | -0.006 | -0.4685 | No | ||
| 56 | KRT33A | 9252 | 12529 | -0.007 | -0.4913 | No | ||
| 57 | LAMC3 | 6241 | 12854 | -0.008 | -0.5074 | No | ||
| 58 | KRT20 | 12410 | 13181 | -0.009 | -0.5235 | No | ||
| 59 | GJA5 | 1929 9017 4779 | 13233 | -0.009 | -0.5246 | No | ||
| 60 | GJB1 | 24276 | 13398 | -0.010 | -0.5317 | No | ||
| 61 | KRT39 | 10701 | 13434 | -0.010 | -0.5319 | No | ||
| 62 | GJB4 | 4782 | 13659 | -0.012 | -0.5420 | No | ||
| 63 | CHAD | 20702 | 13873 | -0.013 | -0.5513 | No | ||
| 64 | LAMC2 | 4983 9266 13806 3975 | 14094 | -0.014 | -0.5607 | No | ||
| 65 | TNXB | 1557 8174 878 8175 | 14366 | -0.016 | -0.5726 | No | ||
| 66 | LAMA3 | 23620 2009 | 15587 | -0.034 | -0.6326 | No | ||
| 67 | ITGA6 | 4930 | 15653 | -0.036 | -0.6300 | No | ||
| 68 | DSG4 | 9262 23616 | 15773 | -0.039 | -0.6298 | No | ||
| 69 | LMNA | 4999 15290 1778 9280 | 16070 | -0.050 | -0.6373 | No | ||
| 70 | LMNB2 | 19685 | 16403 | -0.065 | -0.6442 | No | ||
| 71 | LAMB2 | 9265 | 16687 | -0.081 | -0.6457 | No | ||
| 72 | THBS2 | 5749 | 17483 | -0.151 | -0.6629 | Yes | ||
| 73 | LMNB1 | 23549 1943 | 17490 | -0.152 | -0.6374 | Yes | ||
| 74 | LAMA4 | 20054 | 17596 | -0.167 | -0.6147 | Yes | ||
| 75 | GJA1 | 20023 | 17606 | -0.169 | -0.5865 | Yes | ||
| 76 | VIM | 80 | 18145 | -0.288 | -0.5666 | Yes | ||
| 77 | ACTB | 8534 337 337 338 | 18610 | -3.484 | 0.0003 | Yes |