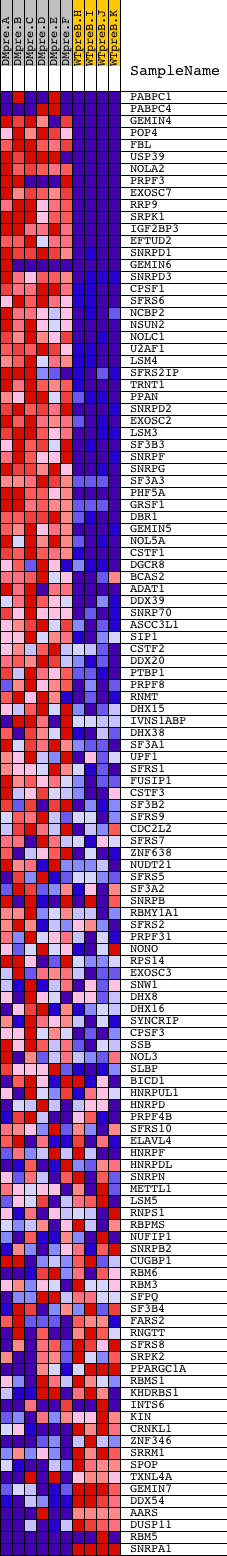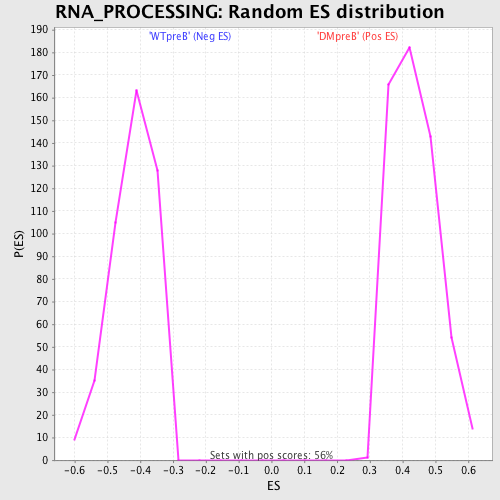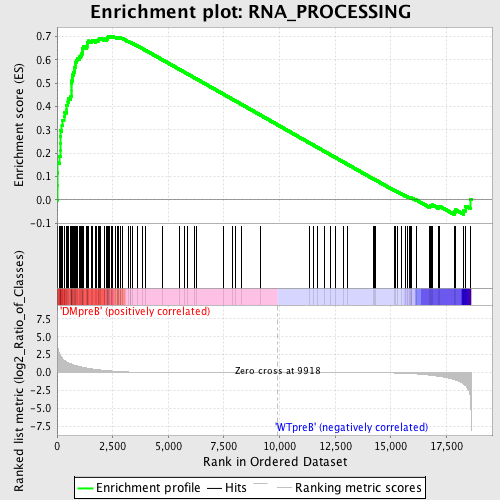
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | 0.701059 |
| Normalized Enrichment Score (NES) | 1.6094522 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.039630834 |
| FWER p-Value | 0.311 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PABPC1 | 5219 9522 9523 23572 | 7 | 5.273 | 0.0617 | Yes | ||
| 2 | PABPC4 | 10510 2418 2548 | 11 | 4.632 | 0.1160 | Yes | ||
| 3 | GEMIN4 | 6592 6591 | 31 | 3.857 | 0.1603 | Yes | ||
| 4 | POP4 | 7261 | 118 | 2.547 | 0.1857 | Yes | ||
| 5 | FBL | 8955 | 142 | 2.450 | 0.2132 | Yes | ||
| 6 | USP39 | 1116 1083 11373 | 147 | 2.443 | 0.2418 | Yes | ||
| 7 | NOLA2 | 11972 | 152 | 2.423 | 0.2701 | Yes | ||
| 8 | PRPF3 | 12899 12898 1877 7703 | 162 | 2.366 | 0.2974 | Yes | ||
| 9 | EXOSC7 | 19256 | 218 | 2.074 | 0.3188 | Yes | ||
| 10 | RRP9 | 19328 | 231 | 2.041 | 0.3422 | Yes | ||
| 11 | SRPK1 | 23049 | 310 | 1.742 | 0.3585 | Yes | ||
| 12 | IGF2BP3 | 4699 8932 | 344 | 1.666 | 0.3763 | Yes | ||
| 13 | EFTUD2 | 1219 20203 | 431 | 1.483 | 0.3891 | Yes | ||
| 14 | SNRPD1 | 23622 | 435 | 1.475 | 0.4063 | Yes | ||
| 15 | GEMIN6 | 12492 7415 | 487 | 1.391 | 0.4199 | Yes | ||
| 16 | SNRPD3 | 12514 | 506 | 1.373 | 0.4351 | Yes | ||
| 17 | CPSF1 | 22241 | 606 | 1.230 | 0.4442 | Yes | ||
| 18 | SFRS6 | 14751 | 645 | 1.178 | 0.4560 | Yes | ||
| 19 | NCBP2 | 12643 | 646 | 1.178 | 0.4698 | Yes | ||
| 20 | NSUN2 | 21610 | 648 | 1.176 | 0.4836 | Yes | ||
| 21 | NOLC1 | 7704 | 649 | 1.175 | 0.4974 | Yes | ||
| 22 | U2AF1 | 8431 | 664 | 1.165 | 0.5104 | Yes | ||
| 23 | LSM4 | 18583 | 695 | 1.122 | 0.5220 | Yes | ||
| 24 | SFRS2IP | 7794 13009 | 699 | 1.114 | 0.5349 | Yes | ||
| 25 | TRNT1 | 17344 | 735 | 1.075 | 0.5457 | Yes | ||
| 26 | PPAN | 19546 | 779 | 1.031 | 0.5555 | Yes | ||
| 27 | SNRPD2 | 8412 | 784 | 1.029 | 0.5674 | Yes | ||
| 28 | EXOSC2 | 15044 | 820 | 1.004 | 0.5773 | Yes | ||
| 29 | LSM3 | 12565 | 827 | 0.998 | 0.5887 | Yes | ||
| 30 | SF3B3 | 18746 | 858 | 0.955 | 0.5983 | Yes | ||
| 31 | SNRPF | 7645 | 897 | 0.925 | 0.6071 | Yes | ||
| 32 | SNRPG | 12622 | 991 | 0.845 | 0.6121 | Yes | ||
| 33 | SF3A3 | 16091 | 1070 | 0.793 | 0.6172 | Yes | ||
| 34 | PHF5A | 22194 | 1098 | 0.775 | 0.6248 | Yes | ||
| 35 | GRSF1 | 16487 | 1122 | 0.759 | 0.6325 | Yes | ||
| 36 | DBR1 | 19341 | 1138 | 0.750 | 0.6405 | Yes | ||
| 37 | GEMIN5 | 20439 | 1141 | 0.746 | 0.6492 | Yes | ||
| 38 | NOL5A | 12474 | 1181 | 0.723 | 0.6556 | Yes | ||
| 39 | CSTF1 | 12515 | 1305 | 0.641 | 0.6565 | Yes | ||
| 40 | DGCR8 | 22644 | 1345 | 0.620 | 0.6617 | Yes | ||
| 41 | BCAS2 | 12662 | 1347 | 0.620 | 0.6689 | Yes | ||
| 42 | ADAT1 | 11400 6633 | 1362 | 0.613 | 0.6753 | Yes | ||
| 43 | DDX39 | 18551 | 1399 | 0.591 | 0.6803 | Yes | ||
| 44 | SNRP70 | 1186 | 1537 | 0.531 | 0.6792 | Yes | ||
| 45 | ASCC3L1 | 14867 | 1576 | 0.512 | 0.6832 | Yes | ||
| 46 | SIP1 | 21263 | 1744 | 0.440 | 0.6793 | Yes | ||
| 47 | CSTF2 | 24257 | 1764 | 0.432 | 0.6834 | Yes | ||
| 48 | DDX20 | 15213 | 1844 | 0.403 | 0.6838 | Yes | ||
| 49 | PTBP1 | 5303 | 1875 | 0.392 | 0.6868 | Yes | ||
| 50 | PRPF8 | 20780 1371 | 1916 | 0.379 | 0.6891 | Yes | ||
| 51 | RNMT | 7501 | 1964 | 0.365 | 0.6909 | Yes | ||
| 52 | DHX15 | 8842 | 2119 | 0.313 | 0.6862 | Yes | ||
| 53 | IVNS1ABP | 4384 4050 | 2138 | 0.308 | 0.6889 | Yes | ||
| 54 | DHX38 | 48 | 2236 | 0.279 | 0.6869 | Yes | ||
| 55 | SF3A1 | 7450 | 2253 | 0.275 | 0.6893 | Yes | ||
| 56 | UPF1 | 9718 18855 | 2259 | 0.274 | 0.6922 | Yes | ||
| 57 | SFRS1 | 8492 | 2269 | 0.270 | 0.6949 | Yes | ||
| 58 | FUSIP1 | 4715 16036 | 2287 | 0.266 | 0.6971 | Yes | ||
| 59 | CSTF3 | 10449 6002 | 2313 | 0.259 | 0.6988 | Yes | ||
| 60 | SF3B2 | 23974 | 2363 | 0.248 | 0.6991 | Yes | ||
| 61 | SFRS9 | 16731 | 2448 | 0.225 | 0.6972 | Yes | ||
| 62 | CDC2L2 | 15964 2419 | 2459 | 0.222 | 0.6993 | Yes | ||
| 63 | SFRS7 | 22889 | 2474 | 0.218 | 0.7011 | Yes | ||
| 64 | ZNF638 | 9476 | 2613 | 0.187 | 0.6958 | No | ||
| 65 | NUDT21 | 12665 | 2696 | 0.167 | 0.6933 | No | ||
| 66 | SFRS5 | 9808 2062 | 2710 | 0.165 | 0.6946 | No | ||
| 67 | SF3A2 | 19938 | 2749 | 0.155 | 0.6943 | No | ||
| 68 | SNRPB | 9842 5469 2736 | 2756 | 0.153 | 0.6958 | No | ||
| 69 | RBMY1A1 | 5368 | 2837 | 0.137 | 0.6931 | No | ||
| 70 | SFRS2 | 9807 20136 | 2845 | 0.136 | 0.6943 | No | ||
| 71 | PRPF31 | 7594 | 2923 | 0.123 | 0.6916 | No | ||
| 72 | NONO | 11993 | 3209 | 0.085 | 0.6772 | No | ||
| 73 | RPS14 | 9751 | 3282 | 0.079 | 0.6742 | No | ||
| 74 | EXOSC3 | 15890 | 3304 | 0.077 | 0.6740 | No | ||
| 75 | SNW1 | 7282 | 3405 | 0.068 | 0.6694 | No | ||
| 76 | DHX8 | 20649 | 3618 | 0.057 | 0.6586 | No | ||
| 77 | DHX16 | 23247 | 3839 | 0.046 | 0.6472 | No | ||
| 78 | SYNCRIP | 3078 3035 3107 | 3969 | 0.042 | 0.6408 | No | ||
| 79 | CPSF3 | 7038 | 4738 | 0.024 | 0.5995 | No | ||
| 80 | SSB | 907 14998 | 5509 | 0.016 | 0.5581 | No | ||
| 81 | NOL3 | 18496 915 921 | 5720 | 0.014 | 0.5469 | No | ||
| 82 | SLBP | 9822 | 5863 | 0.013 | 0.5394 | No | ||
| 83 | BICD1 | 8657 4447 | 6182 | 0.011 | 0.5223 | No | ||
| 84 | HNRPUL1 | 6118 17923 1608 10577 1345 | 6268 | 0.011 | 0.5178 | No | ||
| 85 | HNRPD | 8645 | 7484 | 0.006 | 0.4522 | No | ||
| 86 | PRPF4B | 3208 5295 | 7903 | 0.005 | 0.4297 | No | ||
| 87 | SFRS10 | 9818 1722 5441 | 8003 | 0.005 | 0.4244 | No | ||
| 88 | ELAVL4 | 15805 4889 9137 | 8277 | 0.004 | 0.4097 | No | ||
| 89 | HNRPF | 13607 | 9135 | 0.002 | 0.3633 | No | ||
| 90 | HNRPDL | 11947 3551 6954 | 11341 | -0.004 | 0.2442 | No | ||
| 91 | SNRPN | 1715 9844 5471 | 11506 | -0.004 | 0.2353 | No | ||
| 92 | METTL1 | 19860 3400 3404 | 11714 | -0.005 | 0.2242 | No | ||
| 93 | LSM5 | 7286 | 12026 | -0.006 | 0.2075 | No | ||
| 94 | RNPS1 | 9730 23361 | 12295 | -0.007 | 0.1930 | No | ||
| 95 | RBPMS | 5369 | 12529 | -0.008 | 0.1805 | No | ||
| 96 | NUFIP1 | 21955 | 12876 | -0.009 | 0.1619 | No | ||
| 97 | SNRPB2 | 5470 | 13065 | -0.010 | 0.1519 | No | ||
| 98 | CUGBP1 | 2805 8819 4576 2924 | 14212 | -0.023 | 0.0902 | No | ||
| 99 | RBM6 | 9707 3095 3063 2995 | 14260 | -0.024 | 0.0879 | No | ||
| 100 | RBM3 | 5366 2625 | 14306 | -0.025 | 0.0858 | No | ||
| 101 | SFPQ | 12936 | 15172 | -0.054 | 0.0397 | No | ||
| 102 | SF3B4 | 22269 | 15231 | -0.058 | 0.0372 | No | ||
| 103 | FARS2 | 21666 | 15310 | -0.063 | 0.0337 | No | ||
| 104 | RNGTT | 10769 2354 6272 | 15467 | -0.074 | 0.0262 | No | ||
| 105 | SFRS8 | 10543 6089 | 15641 | -0.090 | 0.0179 | No | ||
| 106 | SRPK2 | 5513 | 15728 | -0.101 | 0.0144 | No | ||
| 107 | PPARGC1A | 16533 | 15834 | -0.119 | 0.0101 | No | ||
| 108 | RBMS1 | 14580 | 15872 | -0.124 | 0.0096 | No | ||
| 109 | KHDRBS1 | 5405 9778 | 15928 | -0.131 | 0.0082 | No | ||
| 110 | INTS6 | 5184 126 | 15932 | -0.132 | 0.0096 | No | ||
| 111 | KIN | 15120 | 16140 | -0.177 | 0.0004 | No | ||
| 112 | CRNKL1 | 14407 | 16745 | -0.354 | -0.0280 | No | ||
| 113 | ZNF346 | 3163 6517 3193 | 16770 | -0.364 | -0.0251 | No | ||
| 114 | SRRM1 | 6958 | 16840 | -0.383 | -0.0243 | No | ||
| 115 | SPOP | 5498 | 16868 | -0.393 | -0.0211 | No | ||
| 116 | TXNL4A | 6567 11329 | 17135 | -0.504 | -0.0296 | No | ||
| 117 | GEMIN7 | 17944 | 17201 | -0.526 | -0.0269 | No | ||
| 118 | DDX54 | 7778 | 17851 | -0.960 | -0.0507 | No | ||
| 119 | AARS | 10630 6151 | 17922 | -1.032 | -0.0423 | No | ||
| 120 | DUSP11 | 17086 7785 | 18283 | -1.597 | -0.0430 | No | ||
| 121 | RBM5 | 13507 3016 | 18365 | -1.822 | -0.0260 | No | ||
| 122 | SNRPA1 | 1373 18216 | 18569 | -3.355 | 0.0025 | No |