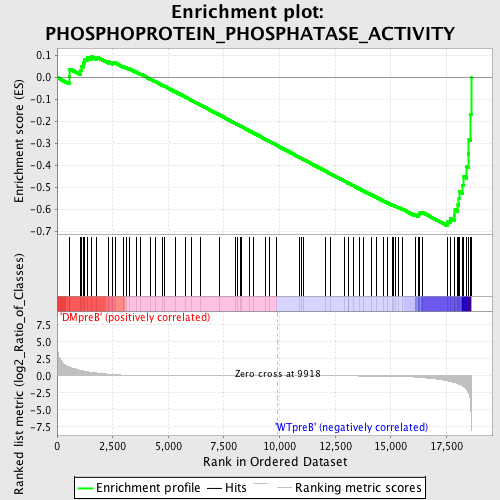
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | PHOSPHOPROTEIN_PHOSPHATASE_ACTIVITY |
| Enrichment Score (ES) | -0.67564064 |
| Normalized Enrichment Score (NES) | -1.4947801 |
| Nominal p-value | 0.008602151 |
| FDR q-value | 0.42057237 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM50 | 17911 | 538 | 1.326 | 0.0058 | No | ||
| 2 | PTPLA | 11403 6636 14680 | 567 | 1.294 | 0.0382 | No | ||
| 3 | PPP5C | 17952 | 1049 | 0.806 | 0.0334 | No | ||
| 4 | CYCS | 8821 | 1104 | 0.771 | 0.0507 | No | ||
| 5 | PPM1G | 16571 | 1167 | 0.734 | 0.0667 | No | ||
| 6 | PPP2CB | 18636 | 1225 | 0.700 | 0.0819 | No | ||
| 7 | PPM1A | 5279 9607 | 1352 | 0.616 | 0.0913 | No | ||
| 8 | PTP4A2 | 9659 2463 2518 5324 | 1562 | 0.520 | 0.0937 | No | ||
| 9 | PTPRA | 14844 | 1791 | 0.423 | 0.0925 | No | ||
| 10 | DLG7 | 21858 | 2294 | 0.264 | 0.0723 | No | ||
| 11 | PTPN7 | 11500 | 2509 | 0.210 | 0.0663 | No | ||
| 12 | CTDP1 | 23399 1969 | 2605 | 0.191 | 0.0662 | No | ||
| 13 | PTPN5 | 17817 | 2979 | 0.114 | 0.0491 | No | ||
| 14 | DLG1 | 22793 1672 | 3114 | 0.096 | 0.0444 | No | ||
| 15 | PPP1R3C | 23688 | 3231 | 0.083 | 0.0403 | No | ||
| 16 | PTPN21 | 21009 | 3566 | 0.059 | 0.0238 | No | ||
| 17 | PTPRS | 1507 1565 1602 22928 | 3746 | 0.050 | 0.0155 | No | ||
| 18 | PTPRN | 13914 | 4213 | 0.035 | -0.0087 | No | ||
| 19 | DUSP3 | 1358 13022 | 4409 | 0.030 | -0.0185 | No | ||
| 20 | DUSP7 | 10661 | 4741 | 0.024 | -0.0357 | No | ||
| 21 | PTPRU | 15738 | 4827 | 0.023 | -0.0397 | No | ||
| 22 | PTPRR | 3436 19876 | 5299 | 0.018 | -0.0646 | No | ||
| 23 | TPTE | 3875 3866 10615 3777 | 5763 | 0.014 | -0.0892 | No | ||
| 24 | EPM2A | 20090 | 6052 | 0.012 | -0.1044 | No | ||
| 25 | PTPRG | 22097 3151 | 6448 | 0.010 | -0.1255 | No | ||
| 26 | PTPRM | 22904 | 7318 | 0.007 | -0.1721 | No | ||
| 27 | PTPRK | 5332 20066 | 8038 | 0.005 | -0.2108 | No | ||
| 28 | PTPN3 | 9661 | 8117 | 0.004 | -0.2149 | No | ||
| 29 | PTPRD | 11554 5330 | 8261 | 0.004 | -0.2225 | No | ||
| 30 | PTPN2 | 23414 | 8307 | 0.004 | -0.2248 | No | ||
| 31 | DUSP12 | 13765 | 8662 | 0.003 | -0.2438 | No | ||
| 32 | PTPRF | 5331 2545 | 8807 | 0.003 | -0.2515 | No | ||
| 33 | PTPN11 | 5326 16391 9660 | 9360 | 0.001 | -0.2813 | No | ||
| 34 | PTPRN2 | 9666 | 9539 | 0.001 | -0.2909 | No | ||
| 35 | DUSP13 | 21904 | 9877 | 0.000 | -0.3090 | No | ||
| 36 | PTPRO | 1093 17256 | 10877 | -0.002 | -0.3628 | No | ||
| 37 | PTPN4 | 13856 | 10985 | -0.003 | -0.3685 | No | ||
| 38 | PPEF2 | 16481 | 11091 | -0.003 | -0.3741 | No | ||
| 39 | PTPRB | 19875 | 12069 | -0.006 | -0.4267 | No | ||
| 40 | MTMR7 | 12032 | 12277 | -0.007 | -0.4377 | No | ||
| 41 | PTPRH | 11861 | 12912 | -0.010 | -0.4716 | No | ||
| 42 | CDKN3 | 16965 | 13076 | -0.011 | -0.4801 | No | ||
| 43 | PPP2R2A | 3222 21774 | 13095 | -0.011 | -0.4808 | No | ||
| 44 | CILP | 19404 | 13335 | -0.012 | -0.4934 | No | ||
| 45 | PTPN14 | 14012 | 13582 | -0.014 | -0.5063 | No | ||
| 46 | PTPRT | 5333 | 13774 | -0.016 | -0.5161 | No | ||
| 47 | PTPN12 | 16597 3502 3586 3665 | 14152 | -0.022 | -0.5359 | No | ||
| 48 | PPAP2C | 6938 | 14375 | -0.026 | -0.5472 | No | ||
| 49 | DUSP4 | 18632 3820 | 14677 | -0.034 | -0.5625 | No | ||
| 50 | SBF1 | 13420 | 14836 | -0.039 | -0.5700 | No | ||
| 51 | CDC14B | 3227 5750 | 15070 | -0.049 | -0.5813 | No | ||
| 52 | DUSP8 | 9493 | 15134 | -0.052 | -0.5833 | No | ||
| 53 | PTPN22 | 1775 15470 | 15214 | -0.057 | -0.5861 | No | ||
| 54 | CDC25B | 14841 | 15324 | -0.063 | -0.5903 | No | ||
| 55 | PTPRC | 5327 9662 | 15526 | -0.080 | -0.5990 | No | ||
| 56 | PTPN9 | 19434 | 16102 | -0.169 | -0.6256 | No | ||
| 57 | PTPN1 | 5325 | 16237 | -0.200 | -0.6276 | No | ||
| 58 | PTPRJ | 9664 | 16260 | -0.207 | -0.6234 | No | ||
| 59 | LCK | 15746 | 16281 | -0.210 | -0.6189 | No | ||
| 60 | MTMR3 | 7908 13193 | 16307 | -0.217 | -0.6146 | No | ||
| 61 | DULLARD | 20816 | 16406 | -0.244 | -0.6135 | No | ||
| 62 | PPP6C | 7491 | 17560 | -0.738 | -0.6563 | Yes | ||
| 63 | PTPN18 | 14279 | 17668 | -0.805 | -0.6409 | Yes | ||
| 64 | PPM1D | 20721 | 17849 | -0.958 | -0.6255 | Yes | ||
| 65 | PPP1CA | 9608 3756 914 909 | 17883 | -0.991 | -0.6013 | Yes | ||
| 66 | PTEN | 5305 | 18017 | -1.128 | -0.5788 | Yes | ||
| 67 | PPAP2B | 7503 16161 | 18063 | -1.194 | -0.5499 | Yes | ||
| 68 | DUSP1 | 23061 | 18067 | -1.196 | -0.5187 | Yes | ||
| 69 | PPM1B | 5280 1567 5281 | 18213 | -1.409 | -0.4896 | Yes | ||
| 70 | DUSP11 | 17086 7785 | 18283 | -1.597 | -0.4514 | Yes | ||
| 71 | DUSP2 | 14865 | 18415 | -1.980 | -0.4065 | Yes | ||
| 72 | PTP4A3 | 22460 2304 | 18476 | -2.353 | -0.3480 | Yes | ||
| 73 | PTPN6 | 17002 | 18512 | -2.552 | -0.2829 | Yes | ||
| 74 | DUSP6 | 19891 3399 | 18594 | -4.601 | -0.1666 | Yes | ||
| 75 | PTPRE | 1662 18039 | 18613 | -6.391 | 0.0002 | Yes |

