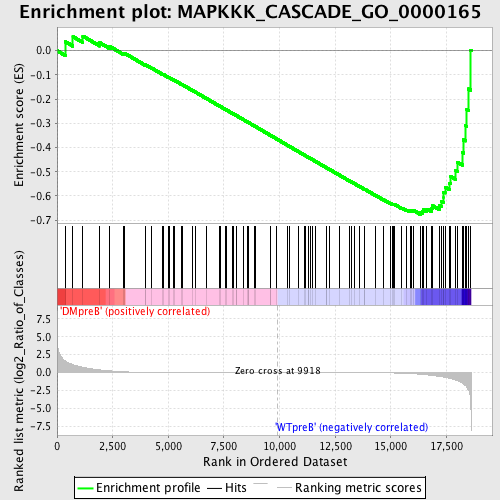
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | MAPKKK_CASCADE_GO_0000165 |
| Enrichment Score (ES) | -0.6765322 |
| Normalized Enrichment Score (NES) | -1.5369203 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.3432743 |
| FWER p-Value | 0.93 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ADRA2C | 16869 | 376 | 1.600 | 0.0369 | No | ||
| 2 | PIK3CB | 19030 | 713 | 1.094 | 0.0579 | No | ||
| 3 | SOD1 | 9846 | 1158 | 0.738 | 0.0603 | No | ||
| 4 | MAPK8 | 6459 | 1918 | 0.379 | 0.0328 | No | ||
| 5 | PKN1 | 11567 6799 | 2353 | 0.251 | 0.0184 | No | ||
| 6 | TRAF7 | 10325 | 2978 | 0.114 | -0.0112 | No | ||
| 7 | DAXX | 23290 | 3036 | 0.104 | -0.0106 | No | ||
| 8 | RGS3 | 2434 2474 6937 11935 2374 2332 2445 2390 | 3960 | 0.042 | -0.0589 | No | ||
| 9 | SHC1 | 9813 9812 5430 | 3966 | 0.042 | -0.0577 | No | ||
| 10 | SPRED2 | 8539 4332 | 4259 | 0.033 | -0.0722 | No | ||
| 11 | CAMKK2 | 9870 | 4758 | 0.024 | -0.0982 | No | ||
| 12 | MDFI | 22950 | 4787 | 0.024 | -0.0989 | No | ||
| 13 | FGF13 | 4720 8963 2627 | 5022 | 0.021 | -0.1108 | No | ||
| 14 | MAP3K9 | 6872 | 5039 | 0.021 | -0.1109 | No | ||
| 15 | MAP3K13 | 22814 | 5229 | 0.019 | -0.1205 | No | ||
| 16 | GRM4 | 332 | 5287 | 0.018 | -0.1229 | No | ||
| 17 | GPS1 | 1392 9943 5550 9944 | 5603 | 0.015 | -0.1394 | No | ||
| 18 | PROK2 | 17057 | 5634 | 0.015 | -0.1404 | No | ||
| 19 | FGFR3 | 8969 3566 | 6077 | 0.012 | -0.1639 | No | ||
| 20 | SPRED1 | 4331 | 6238 | 0.011 | -0.1721 | No | ||
| 21 | ADRA2A | 4355 | 6724 | 0.009 | -0.1980 | No | ||
| 22 | C5AR1 | 17957 | 7292 | 0.007 | -0.2283 | No | ||
| 23 | NRTN | 22923 | 7347 | 0.007 | -0.2310 | No | ||
| 24 | TAOK2 | 10610 3794 | 7578 | 0.006 | -0.2432 | No | ||
| 25 | PAK1 | 9527 | 7592 | 0.006 | -0.2437 | No | ||
| 26 | ADRB3 | 18901 | 7884 | 0.005 | -0.2592 | No | ||
| 27 | ATP6AP2 | 2567 24380 2579 2656 | 7947 | 0.005 | -0.2624 | No | ||
| 28 | MAP3K10 | 3831 17916 | 8050 | 0.004 | -0.2678 | No | ||
| 29 | GNG3 | 23752 | 8072 | 0.004 | -0.2687 | No | ||
| 30 | MAP3K7IP1 | 2193 2171 22419 | 8369 | 0.004 | -0.2846 | No | ||
| 31 | DUSP22 | 466 21680 | 8572 | 0.003 | -0.2954 | No | ||
| 32 | MBIP | 21063 2072 | 8583 | 0.003 | -0.2958 | No | ||
| 33 | MAP2K7 | 6453 | 8614 | 0.003 | -0.2973 | No | ||
| 34 | MAP3K6 | 16054 | 8853 | 0.003 | -0.3101 | No | ||
| 35 | ADORA2B | 20850 | 8923 | 0.002 | -0.3137 | No | ||
| 36 | EDA2R | 6364 | 9593 | 0.001 | -0.3498 | No | ||
| 37 | CHRNA7 | 17808 | 9599 | 0.001 | -0.3500 | No | ||
| 38 | MINK1 | 20802 | 9865 | 0.000 | -0.3643 | No | ||
| 39 | DUSP9 | 24307 | 10373 | -0.001 | -0.3917 | No | ||
| 40 | EPGN | 16798 | 10442 | -0.001 | -0.3953 | No | ||
| 41 | HIPK2 | 17180 | 10830 | -0.002 | -0.4161 | No | ||
| 42 | SCG2 | 5410 14425 | 11138 | -0.003 | -0.4326 | No | ||
| 43 | MAPK8IP3 | 11402 23080 | 11152 | -0.003 | -0.4332 | No | ||
| 44 | GHRL | 1183 17040 | 11284 | -0.003 | -0.4401 | No | ||
| 45 | FPR1 | 23115 | 11300 | -0.003 | -0.4408 | No | ||
| 46 | PLCE1 | 13156 | 11373 | -0.004 | -0.4446 | No | ||
| 47 | MAP3K12 | 6454 11164 | 11464 | -0.004 | -0.4493 | No | ||
| 48 | TDGF1 | 5704 10108 | 11624 | -0.004 | -0.4577 | No | ||
| 49 | TGFA | 17381 | 12092 | -0.006 | -0.4827 | No | ||
| 50 | EGF | 15169 | 12130 | -0.006 | -0.4845 | No | ||
| 51 | RGS4 | 9724 | 12233 | -0.007 | -0.4897 | No | ||
| 52 | DUSP16 | 1004 7699 | 12672 | -0.009 | -0.5131 | No | ||
| 53 | MADD | 2926 14526 | 13136 | -0.011 | -0.5377 | No | ||
| 54 | FGFR1 | 3789 8968 | 13224 | -0.012 | -0.5420 | No | ||
| 55 | MAP3K5 | 20079 11166 | 13358 | -0.013 | -0.5487 | No | ||
| 56 | MAPK10 | 11169 | 13577 | -0.014 | -0.5599 | No | ||
| 57 | MAPK8IP2 | 77 | 13813 | -0.017 | -0.5720 | No | ||
| 58 | MAP3K4 | 23126 | 13828 | -0.017 | -0.5722 | No | ||
| 59 | CD27 | 16994 | 14292 | -0.024 | -0.5963 | No | ||
| 60 | DUSP4 | 18632 3820 | 14677 | -0.034 | -0.6158 | No | ||
| 61 | TPD52L1 | 3420 19793 | 14978 | -0.045 | -0.6304 | No | ||
| 62 | MAP4K5 | 11853 21048 2101 2142 | 15067 | -0.049 | -0.6334 | No | ||
| 63 | DUSP8 | 9493 | 15134 | -0.052 | -0.6351 | No | ||
| 64 | FGF2 | 15608 | 15175 | -0.054 | -0.6353 | No | ||
| 65 | MAP3K2 | 11165 | 15499 | -0.077 | -0.6500 | No | ||
| 66 | NF1 | 5165 | 15715 | -0.100 | -0.6580 | No | ||
| 67 | CARTPT | 21374 | 15862 | -0.123 | -0.6615 | No | ||
| 68 | SH2D3C | 2864 | 15941 | -0.134 | -0.6609 | No | ||
| 69 | MAP4K3 | 22887 | 16032 | -0.154 | -0.6603 | No | ||
| 70 | MAP4K2 | 6457 | 16334 | -0.224 | -0.6685 | Yes | ||
| 71 | EDG5 | 19216 | 16428 | -0.252 | -0.6645 | Yes | ||
| 72 | CD24 | 8711 | 16453 | -0.262 | -0.6565 | Yes | ||
| 73 | MAP4K1 | 18313 | 16615 | -0.312 | -0.6540 | Yes | ||
| 74 | MAP2K4 | 20405 | 16842 | -0.384 | -0.6525 | Yes | ||
| 75 | GADD45G | 21637 | 16864 | -0.392 | -0.6396 | Yes | ||
| 76 | DBNL | 20954 | 17191 | -0.523 | -0.6386 | Yes | ||
| 77 | GPS2 | 12096 | 17275 | -0.564 | -0.6229 | Yes | ||
| 78 | CRKL | 4560 | 17368 | -0.613 | -0.6060 | Yes | ||
| 79 | GADD45B | 19935 | 17372 | -0.615 | -0.5841 | Yes | ||
| 80 | DUSP10 | 4003 14016 | 17441 | -0.668 | -0.5639 | Yes | ||
| 81 | CD81 | 8719 | 17641 | -0.785 | -0.5466 | Yes | ||
| 82 | ADRB2 | 23422 | 17695 | -0.828 | -0.5199 | Yes | ||
| 83 | MAP3K11 | 11163 | 17905 | -1.013 | -0.4949 | Yes | ||
| 84 | MAP3K3 | 20626 | 17998 | -1.103 | -0.4605 | Yes | ||
| 85 | CCM2 | 20950 | 18223 | -1.441 | -0.4211 | Yes | ||
| 86 | MAPKAPK2 | 13838 | 18261 | -1.536 | -0.3682 | Yes | ||
| 87 | MAPK9 | 1233 20903 1383 | 18352 | -1.775 | -0.3096 | Yes | ||
| 88 | DUSP2 | 14865 | 18415 | -1.980 | -0.2422 | Yes | ||
| 89 | CXCR4 | 13844 | 18494 | -2.475 | -0.1579 | Yes | ||
| 90 | DUSP6 | 19891 3399 | 18594 | -4.601 | 0.0012 | Yes |

