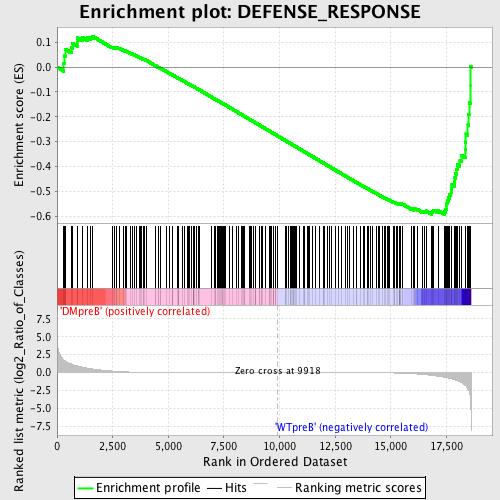
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | DEFENSE_RESPONSE |
| Enrichment Score (ES) | -0.5936042 |
| Normalized Enrichment Score (NES) | -1.5005114 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.44178656 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TPST1 | 10214 | 304 | 1.759 | 0.0154 | No | ||
| 2 | BLNK | 23681 3691 | 325 | 1.702 | 0.0451 | No | ||
| 3 | TARBP2 | 10033 10034 5626 | 377 | 1.600 | 0.0713 | No | ||
| 4 | CYSLTR1 | 24082 | 642 | 1.182 | 0.0784 | No | ||
| 5 | KCNN4 | 18353 | 702 | 1.111 | 0.0954 | No | ||
| 6 | CD48 | 14051 | 906 | 0.918 | 0.1010 | No | ||
| 7 | ABCF1 | 22996 10329 5897 | 909 | 0.917 | 0.1175 | No | ||
| 8 | SCYE1 | 8897 | 1130 | 0.756 | 0.1192 | No | ||
| 9 | CEBPB | 8733 | 1369 | 0.609 | 0.1173 | No | ||
| 10 | PGLYRP1 | 18369 | 1511 | 0.539 | 0.1195 | No | ||
| 11 | ITGB1 | 3872 18411 | 1587 | 0.506 | 0.1246 | No | ||
| 12 | NLRC4 | 11225 | 2468 | 0.219 | 0.0807 | No | ||
| 13 | HP | 3805 18750 | 2565 | 0.198 | 0.0791 | No | ||
| 14 | NFX1 | 2475 | 2650 | 0.179 | 0.0778 | No | ||
| 15 | CAMP | 18990 | 2667 | 0.176 | 0.0801 | No | ||
| 16 | S100A9 | 9774 | 2794 | 0.145 | 0.0759 | No | ||
| 17 | PGLYRP4 | 11810 | 2987 | 0.113 | 0.0675 | No | ||
| 18 | C2 | 23013 | 3052 | 0.102 | 0.0659 | No | ||
| 19 | S100A8 | 15527 | 3112 | 0.096 | 0.0644 | No | ||
| 20 | UMOD | 17660 | 3300 | 0.077 | 0.0556 | No | ||
| 21 | CLEC5A | 6225 | 3375 | 0.070 | 0.0529 | No | ||
| 22 | DMBT1 | 18050 | 3464 | 0.064 | 0.0493 | No | ||
| 23 | NFRKB | 10642 | 3558 | 0.060 | 0.0453 | No | ||
| 24 | NCF2 | 14098 5151 9446 | 3691 | 0.053 | 0.0391 | No | ||
| 25 | LTB4R | 22003 | 3738 | 0.050 | 0.0375 | No | ||
| 26 | CCL22 | 18518 | 3774 | 0.049 | 0.0365 | No | ||
| 27 | CXCL9 | 5099 | 3864 | 0.045 | 0.0325 | No | ||
| 28 | ORM1 | 9516 | 3899 | 0.044 | 0.0314 | No | ||
| 29 | LGALS3BP | 20129 | 3942 | 0.042 | 0.0299 | No | ||
| 30 | KLRG1 | 17014 | 4036 | 0.039 | 0.0256 | No | ||
| 31 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 4399 | 0.030 | 0.0064 | No | ||
| 32 | SFTPD | 21867 | 4407 | 0.030 | 0.0066 | No | ||
| 33 | COLEC12 | 23624 8946 | 4545 | 0.027 | -0.0004 | No | ||
| 34 | ALOX5AP | 16616 | 4554 | 0.027 | -0.0003 | No | ||
| 35 | XCR1 | 18953 | 4651 | 0.026 | -0.0050 | No | ||
| 36 | AGER | 23276 1576 | 4665 | 0.025 | -0.0053 | No | ||
| 37 | IL4 | 9174 | 4896 | 0.022 | -0.0174 | No | ||
| 38 | LY75 | 9313 5026 | 5042 | 0.021 | -0.0249 | No | ||
| 39 | IL1A | 4915 | 5169 | 0.019 | -0.0314 | No | ||
| 40 | PGLYRP2 | 7174 | 5201 | 0.019 | -0.0327 | No | ||
| 41 | ITK | 4934 | 5203 | 0.019 | -0.0324 | No | ||
| 42 | MLF2 | 17283 | 5403 | 0.017 | -0.0429 | No | ||
| 43 | CRTAM | 19160 | 5441 | 0.017 | -0.0447 | No | ||
| 44 | TLR7 | 24004 | 5446 | 0.016 | -0.0446 | No | ||
| 45 | MBL2 | 23886 | 5470 | 0.016 | -0.0455 | No | ||
| 46 | SPN | 452 5493 | 5623 | 0.015 | -0.0535 | No | ||
| 47 | CCL24 | 16342 | 5737 | 0.014 | -0.0594 | No | ||
| 48 | IL8RA | 13925 | 5870 | 0.013 | -0.0663 | No | ||
| 49 | CCR2 | 19250 | 5888 | 0.013 | -0.0670 | No | ||
| 50 | BNIP3 | 17581 | 5943 | 0.013 | -0.0697 | No | ||
| 51 | IL5 | 20884 10220 | 6024 | 0.012 | -0.0738 | No | ||
| 52 | NOX4 | 18196 | 6116 | 0.012 | -0.0786 | No | ||
| 53 | CCL5 | 20320 | 6124 | 0.012 | -0.0787 | No | ||
| 54 | TLR3 | 18884 | 6132 | 0.012 | -0.0789 | No | ||
| 55 | DEFB103A | 18665 | 6134 | 0.012 | -0.0788 | No | ||
| 56 | AIF1 | 1582 23005 | 6157 | 0.012 | -0.0797 | No | ||
| 57 | SELE | 14074 | 6269 | 0.011 | -0.0856 | No | ||
| 58 | DEFB1 | 18663 | 6351 | 0.011 | -0.0898 | No | ||
| 59 | CBARA1 | 20015 | 6367 | 0.010 | -0.0904 | No | ||
| 60 | AOC3 | 8595 | 6415 | 0.010 | -0.0928 | No | ||
| 61 | CDO1 | 8731 | 6960 | 0.008 | -0.1222 | No | ||
| 62 | OR2H2 | 17694 | 7089 | 0.008 | -0.1290 | No | ||
| 63 | TFF3 | 23038 | 7102 | 0.007 | -0.1295 | No | ||
| 64 | GHSR | 1810 15625 | 7131 | 0.007 | -0.1309 | No | ||
| 65 | CD160 | 12023 | 7203 | 0.007 | -0.1346 | No | ||
| 66 | IL13 | 20461 | 7272 | 0.007 | -0.1382 | No | ||
| 67 | C5AR1 | 17957 | 7292 | 0.007 | -0.1391 | No | ||
| 68 | C3AR1 | 8668 | 7323 | 0.007 | -0.1406 | No | ||
| 69 | CCR9 | 8753 | 7393 | 0.006 | -0.1443 | No | ||
| 70 | ANKRD1 | 23689 | 7411 | 0.006 | -0.1451 | No | ||
| 71 | GATA3 | 9004 | 7432 | 0.006 | -0.1460 | No | ||
| 72 | IFNA2 | 9149 | 7468 | 0.006 | -0.1478 | No | ||
| 73 | STAB2 | 19656 | 7537 | 0.006 | -0.1514 | No | ||
| 74 | IL9 | 21444 | 7564 | 0.006 | -0.1527 | No | ||
| 75 | IL8RB | 8752 4533 | 7746 | 0.005 | -0.1625 | No | ||
| 76 | TNFAIP6 | 10203 | 7886 | 0.005 | -0.1699 | No | ||
| 77 | LALBA | 22141 | 8075 | 0.004 | -0.1801 | No | ||
| 78 | CCL3 | 20317 | 8172 | 0.004 | -0.1852 | No | ||
| 79 | CD40LG | 24330 | 8298 | 0.004 | -0.1919 | No | ||
| 80 | IL1RL2 | 4212 | 8332 | 0.004 | -0.1936 | No | ||
| 81 | CD5L | 15560 | 8388 | 0.004 | -0.1966 | No | ||
| 82 | GPR68 | 21004 | 8430 | 0.003 | -0.1987 | No | ||
| 83 | IFNK | 11837 | 8443 | 0.003 | -0.1993 | No | ||
| 84 | CCR1 | 18951 | 8645 | 0.003 | -0.2102 | No | ||
| 85 | CX3CL1 | 9791 | 8685 | 0.003 | -0.2123 | No | ||
| 86 | FPRL1 | 8982 | 8733 | 0.003 | -0.2148 | No | ||
| 87 | AHSG | 22812 | 8838 | 0.003 | -0.2204 | No | ||
| 88 | LBP | 14760 | 8894 | 0.002 | -0.2233 | No | ||
| 89 | CYBB | 4579 | 8915 | 0.002 | -0.2243 | No | ||
| 90 | ADORA2B | 20850 | 8923 | 0.002 | -0.2247 | No | ||
| 91 | ELF3 | 8894 13824 | 9097 | 0.002 | -0.2340 | No | ||
| 92 | WFDC12 | 14356 | 9102 | 0.002 | -0.2342 | No | ||
| 93 | PTX3 | 9668 15575 | 9185 | 0.002 | -0.2387 | No | ||
| 94 | IL28RA | 10823 16038 | 9193 | 0.002 | -0.2390 | No | ||
| 95 | EDG3 | 21639 | 9227 | 0.002 | -0.2408 | No | ||
| 96 | PLA2G7 | 23233 | 9373 | 0.001 | -0.2486 | No | ||
| 97 | CXCL10 | 16477 | 9536 | 0.001 | -0.2574 | No | ||
| 98 | ADORA2A | 8557 | 9558 | 0.001 | -0.2585 | No | ||
| 99 | CHRNA7 | 17808 | 9599 | 0.001 | -0.2607 | No | ||
| 100 | AOAH | 11296 | 9635 | 0.001 | -0.2626 | No | ||
| 101 | CCL11 | 20742 | 9713 | 0.000 | -0.2668 | No | ||
| 102 | PRF1 | 20011 | 9734 | 0.000 | -0.2678 | No | ||
| 103 | SOCS6 | 12042 7044 | 9738 | 0.000 | -0.2680 | No | ||
| 104 | IL20 | 13840 | 9807 | 0.000 | -0.2717 | No | ||
| 105 | ORM2 | 16193 | 9837 | 0.000 | -0.2733 | No | ||
| 106 | STAB1 | 21892 | 9894 | 0.000 | -0.2763 | No | ||
| 107 | NCR1 | 18409 | 10245 | -0.001 | -0.2953 | No | ||
| 108 | CXCL11 | 16478 | 10253 | -0.001 | -0.2957 | No | ||
| 109 | CCL4 | 1362 20730 | 10317 | -0.001 | -0.2991 | No | ||
| 110 | TLR8 | 9308 | 10380 | -0.001 | -0.3024 | No | ||
| 111 | IL12B | 20918 | 10475 | -0.001 | -0.3075 | No | ||
| 112 | TACR1 | 1082 17403 | 10505 | -0.001 | -0.3091 | No | ||
| 113 | CCR5 | 8754 4534 | 10515 | -0.001 | -0.3095 | No | ||
| 114 | PLA2G2E | 16016 | 10592 | -0.002 | -0.3136 | No | ||
| 115 | ADORA3 | 4352 | 10635 | -0.002 | -0.3159 | No | ||
| 116 | EREG | 4679 16797 | 10638 | -0.002 | -0.3159 | No | ||
| 117 | APOA4 | 4401 | 10647 | -0.002 | -0.3164 | No | ||
| 118 | APCS | 13748 | 10725 | -0.002 | -0.3205 | No | ||
| 119 | NFATC4 | 22002 | 10757 | -0.002 | -0.3221 | No | ||
| 120 | ANXA1 | 9282 | 10887 | -0.002 | -0.3291 | No | ||
| 121 | IL17C | 18438 | 11062 | -0.003 | -0.3385 | No | ||
| 122 | DCDC2 | 9703 | 11101 | -0.003 | -0.3405 | No | ||
| 123 | SCG2 | 5410 14425 | 11138 | -0.003 | -0.3424 | No | ||
| 124 | PGLYRP3 | 10812 | 11245 | -0.003 | -0.3481 | No | ||
| 125 | GHRL | 1183 17040 | 11284 | -0.003 | -0.3501 | No | ||
| 126 | KLRC3 | 16975 | 11350 | -0.004 | -0.3536 | No | ||
| 127 | CXCL2 | 9790 | 11462 | -0.004 | -0.3596 | No | ||
| 128 | CADM1 | 7057 | 11628 | -0.004 | -0.3684 | No | ||
| 129 | TIAL1 | 996 5753 | 11789 | -0.005 | -0.3771 | No | ||
| 130 | F11R | 9199 | 11952 | -0.005 | -0.3858 | No | ||
| 131 | AOX1 | 14246 4090 | 11968 | -0.005 | -0.3865 | No | ||
| 132 | MST1R | 19318 3106 | 12012 | -0.006 | -0.3887 | No | ||
| 133 | CCR3 | 19251 | 12158 | -0.006 | -0.3965 | No | ||
| 134 | INHBB | 13858 | 12235 | -0.007 | -0.4005 | No | ||
| 135 | IL18RAP | 14262 | 12314 | -0.007 | -0.4046 | No | ||
| 136 | RIPK2 | 2528 15935 | 12516 | -0.008 | -0.4154 | No | ||
| 137 | MEFV | 22686 | 12669 | -0.009 | -0.4235 | No | ||
| 138 | CRP | 8793 | 12766 | -0.009 | -0.4285 | No | ||
| 139 | IL17RB | 21898 | 12978 | -0.010 | -0.4398 | No | ||
| 140 | CX3CR1 | 18965 | 13062 | -0.010 | -0.4441 | No | ||
| 141 | FOSL1 | 23779 | 13138 | -0.011 | -0.4480 | No | ||
| 142 | FOXN1 | 20337 | 13332 | -0.012 | -0.4583 | No | ||
| 143 | HRH1 | 17321 | 13459 | -0.013 | -0.4649 | No | ||
| 144 | ALOX15 | 20370 | 13467 | -0.013 | -0.4650 | No | ||
| 145 | SPACA3 | 13680 | 13617 | -0.015 | -0.4728 | No | ||
| 146 | ADORA1 | 13831 | 13767 | -0.016 | -0.4806 | No | ||
| 147 | MX1 | 9432 22529 | 13825 | -0.017 | -0.4834 | No | ||
| 148 | TNIP1 | 12192 20450 1320 | 13932 | -0.018 | -0.4889 | No | ||
| 149 | KLRC2 | 9243 | 13951 | -0.018 | -0.4895 | No | ||
| 150 | MGLL | 17368 1145 | 14017 | -0.019 | -0.4927 | No | ||
| 151 | CLEC1B | 17270 1080 | 14074 | -0.020 | -0.4954 | No | ||
| 152 | IL1RAP | 1653 1676 4917 1748 1712 9173 | 14177 | -0.022 | -0.5005 | No | ||
| 153 | CXCL1 | 9044 | 14336 | -0.025 | -0.5086 | No | ||
| 154 | CFHR1 | 11926 13815 | 14423 | -0.027 | -0.5128 | No | ||
| 155 | NCF1 | 16354 | 14507 | -0.029 | -0.5168 | No | ||
| 156 | NMI | 14590 | 14632 | -0.032 | -0.5229 | No | ||
| 157 | PLA2G2D | 16018 | 14737 | -0.036 | -0.5279 | No | ||
| 158 | FOS | 21202 | 14780 | -0.038 | -0.5295 | No | ||
| 159 | CCR4 | 18978 | 14828 | -0.039 | -0.5314 | No | ||
| 160 | NLRP3 | 20862 1262 | 14874 | -0.041 | -0.5331 | No | ||
| 161 | LYST | 9325 5040 | 14923 | -0.042 | -0.5349 | No | ||
| 162 | CSF3R | 8804 8803 2326 4565 | 15129 | -0.052 | -0.5451 | No | ||
| 163 | CHST2 | 7032 | 15145 | -0.053 | -0.5450 | No | ||
| 164 | RSAD2 | 21094 | 15164 | -0.054 | -0.5450 | No | ||
| 165 | DEFB127 | 622 | 15247 | -0.059 | -0.5484 | No | ||
| 166 | MX2 | 9433 | 15305 | -0.062 | -0.5503 | No | ||
| 167 | KNG1 | 9244 22809 | 15370 | -0.066 | -0.5526 | No | ||
| 168 | NOD1 | 17141 | 15383 | -0.068 | -0.5520 | No | ||
| 169 | CCR6 | 1505 23390 | 15385 | -0.068 | -0.5509 | No | ||
| 170 | TNFRSF1A | 1181 10206 | 15409 | -0.069 | -0.5509 | No | ||
| 171 | TCIRG1 | 3700 3752 23951 | 15423 | -0.070 | -0.5503 | No | ||
| 172 | CD40 | 10207 2961 2968 5783 | 15430 | -0.071 | -0.5493 | No | ||
| 173 | HDAC9 | 21084 2131 | 15520 | -0.080 | -0.5527 | No | ||
| 174 | PTPRC | 5327 9662 | 15526 | -0.080 | -0.5515 | No | ||
| 175 | NOD2 | 6384 | 15923 | -0.131 | -0.5707 | No | ||
| 176 | BECN1 | 7077 | 16018 | -0.150 | -0.5731 | No | ||
| 177 | PARP4 | 21999 | 16064 | -0.161 | -0.5726 | No | ||
| 178 | CAMLG | 21627 | 16074 | -0.163 | -0.5701 | No | ||
| 179 | TYROBP | 18302 | 16190 | -0.189 | -0.5730 | No | ||
| 180 | LY96 | 14292 | 16426 | -0.252 | -0.5812 | No | ||
| 181 | ZNF148 | 5949 | 16533 | -0.285 | -0.5818 | No | ||
| 182 | RAC1 | 16302 | 16592 | -0.304 | -0.5794 | No | ||
| 183 | IL10RB | 9169 | 16849 | -0.386 | -0.5863 | Yes | ||
| 184 | NFE2L1 | 9457 | 16858 | -0.388 | -0.5797 | Yes | ||
| 185 | VPS45 | 15242 | 16928 | -0.416 | -0.5759 | Yes | ||
| 186 | CCR7 | 20254 | 17126 | -0.500 | -0.5776 | Yes | ||
| 187 | PRDX5 | 7053 3732 | 17422 | -0.653 | -0.5818 | Yes | ||
| 188 | CD19 | 17640 | 17470 | -0.683 | -0.5719 | Yes | ||
| 189 | IRAK2 | 17324 | 17494 | -0.701 | -0.5605 | Yes | ||
| 190 | CD84 | 14049 | 17497 | -0.703 | -0.5479 | Yes | ||
| 191 | NFATC3 | 5169 | 17539 | -0.726 | -0.5369 | Yes | ||
| 192 | NFKB1 | 15160 | 17596 | -0.757 | -0.5262 | Yes | ||
| 193 | CD81 | 8719 | 17641 | -0.785 | -0.5144 | Yes | ||
| 194 | BNIP3L | 21775 | 17704 | -0.839 | -0.5026 | Yes | ||
| 195 | RELA | 23783 | 17734 | -0.859 | -0.4886 | Yes | ||
| 196 | CD83 | 21653 | 17743 | -0.864 | -0.4733 | Yes | ||
| 197 | HDAC5 | 1480 20205 | 17852 | -0.961 | -0.4618 | Yes | ||
| 198 | BCL10 | 15397 | 17878 | -0.984 | -0.4453 | Yes | ||
| 199 | HDAC7A | 22147 | 17916 | -1.026 | -0.4287 | Yes | ||
| 200 | WAS | 24193 | 17956 | -1.065 | -0.4115 | Yes | ||
| 201 | LSP1 | 18003 | 17990 | -1.095 | -0.3935 | Yes | ||
| 202 | VEZF1 | 20709 | 18107 | -1.250 | -0.3771 | Yes | ||
| 203 | MPO | 5116 20710 1324 | 18180 | -1.357 | -0.3565 | Yes | ||
| 204 | PTPRCAP | 23767 | 18339 | -1.751 | -0.3333 | Yes | ||
| 205 | IL12A | 4913 | 18353 | -1.783 | -0.3017 | Yes | ||
| 206 | TGFB1 | 18332 | 18378 | -1.858 | -0.2693 | Yes | ||
| 207 | TLR6 | 215 16528 | 18467 | -2.294 | -0.2325 | Yes | ||
| 208 | CXCR4 | 13844 | 18494 | -2.475 | -0.1891 | Yes | ||
| 209 | CFP | 24174 | 18516 | -2.577 | -0.1435 | Yes | ||
| 210 | RNASE6 | 13439 | 18585 | -3.993 | -0.0748 | Yes | ||
| 211 | CD97 | 18822 3874 | 18587 | -4.218 | 0.0016 | Yes |

