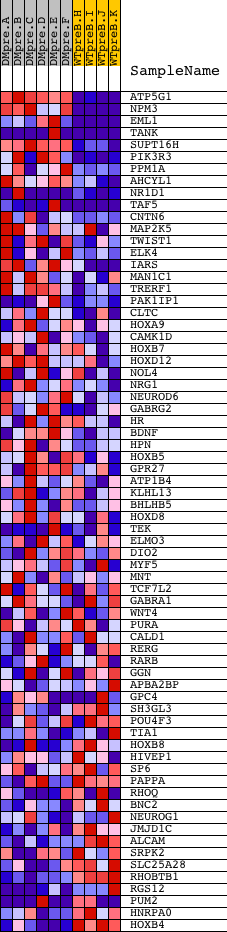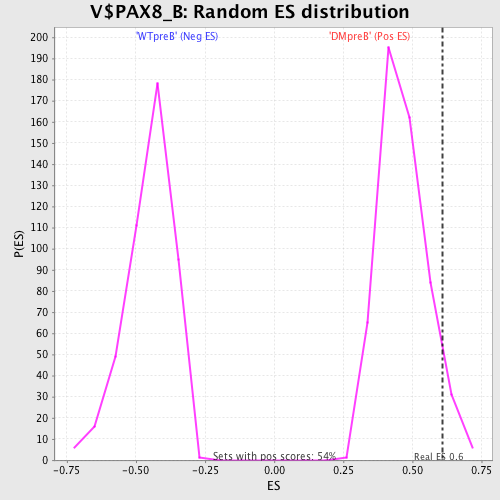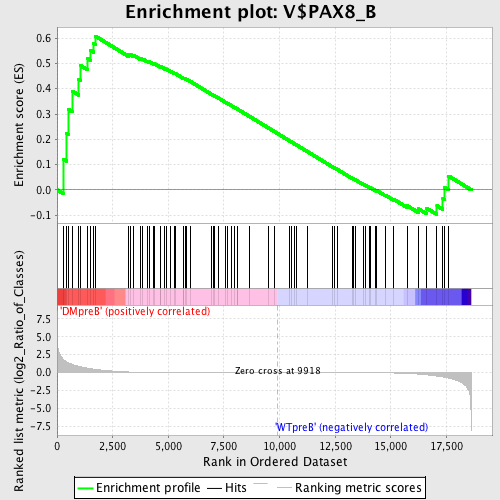
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | V$PAX8_B |
| Enrichment Score (ES) | 0.6078333 |
| Normalized Enrichment Score (NES) | 1.3059117 |
| Nominal p-value | 0.06433824 |
| FDR q-value | 0.7877405 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP5G1 | 8636 | 270 | 1.873 | 0.1219 | Yes | ||
| 2 | NPM3 | 17387 | 428 | 1.487 | 0.2218 | Yes | ||
| 3 | EML1 | 21157 | 501 | 1.377 | 0.3182 | Yes | ||
| 4 | TANK | 2839 15006 2729 | 693 | 1.123 | 0.3897 | Yes | ||
| 5 | SUPT16H | 8541 | 974 | 0.861 | 0.4374 | Yes | ||
| 6 | PIK3R3 | 5248 | 1062 | 0.797 | 0.4908 | Yes | ||
| 7 | PPM1A | 5279 9607 | 1352 | 0.616 | 0.5201 | Yes | ||
| 8 | AHCYL1 | 6037 | 1504 | 0.542 | 0.5514 | Yes | ||
| 9 | NR1D1 | 4650 20258 949 | 1638 | 0.483 | 0.5794 | Yes | ||
| 10 | TAF5 | 23833 5934 3759 | 1719 | 0.449 | 0.6078 | Yes | ||
| 11 | CNTN6 | 17346 | 3205 | 0.085 | 0.5339 | No | ||
| 12 | MAP2K5 | 19088 | 3284 | 0.079 | 0.5355 | No | ||
| 13 | TWIST1 | 21291 | 3422 | 0.067 | 0.5330 | No | ||
| 14 | ELK4 | 8895 4664 3937 | 3729 | 0.051 | 0.5202 | No | ||
| 15 | IARS | 4190 8361 8362 | 3855 | 0.046 | 0.5168 | No | ||
| 16 | MAN1C1 | 10514 6064 15714 | 4061 | 0.039 | 0.5085 | No | ||
| 17 | TRERF1 | 1546 23213 | 4149 | 0.036 | 0.5065 | No | ||
| 18 | PAK1IP1 | 21660 | 4323 | 0.032 | 0.4995 | No | ||
| 19 | CLTC | 14610 | 4355 | 0.031 | 0.5001 | No | ||
| 20 | HOXA9 | 17147 9109 1024 | 4626 | 0.026 | 0.4874 | No | ||
| 21 | CAMK1D | 5977 2948 | 4658 | 0.025 | 0.4876 | No | ||
| 22 | HOXB7 | 666 9110 | 4847 | 0.023 | 0.4791 | No | ||
| 23 | HOXD12 | 9113 | 4899 | 0.022 | 0.4780 | No | ||
| 24 | NOL4 | 2028 11432 | 5092 | 0.020 | 0.4691 | No | ||
| 25 | NRG1 | 3885 | 5293 | 0.018 | 0.4596 | No | ||
| 26 | NEUROD6 | 17138 | 5322 | 0.018 | 0.4594 | No | ||
| 27 | GABRG2 | 1259 4748 8996 1331 | 5685 | 0.014 | 0.4409 | No | ||
| 28 | HR | 3622 9119 | 5702 | 0.014 | 0.4411 | No | ||
| 29 | BDNF | 14926 2797 | 5772 | 0.014 | 0.4384 | No | ||
| 30 | HPN | 17873 4087 | 5828 | 0.013 | 0.4364 | No | ||
| 31 | HOXB5 | 20687 | 5996 | 0.012 | 0.4283 | No | ||
| 32 | GPR27 | 13703 | 6955 | 0.008 | 0.3773 | No | ||
| 33 | ATP1B4 | 12586 | 7032 | 0.008 | 0.3737 | No | ||
| 34 | KLHL13 | 12536 | 7082 | 0.008 | 0.3716 | No | ||
| 35 | BHLHB5 | 15629 | 7249 | 0.007 | 0.3632 | No | ||
| 36 | HOXD8 | 9116 | 7590 | 0.006 | 0.3453 | No | ||
| 37 | TEK | 16175 | 7668 | 0.006 | 0.3415 | No | ||
| 38 | ELMO3 | 10629 18495 | 7850 | 0.005 | 0.3321 | No | ||
| 39 | DIO2 | 21014 2159 | 7975 | 0.005 | 0.3258 | No | ||
| 40 | MYF5 | 19632 | 8086 | 0.004 | 0.3202 | No | ||
| 41 | MNT | 20784 | 8099 | 0.004 | 0.3199 | No | ||
| 42 | TCF7L2 | 10048 5646 | 8665 | 0.003 | 0.2896 | No | ||
| 43 | GABRA1 | 20492 | 9494 | 0.001 | 0.2450 | No | ||
| 44 | WNT4 | 16025 | 9763 | 0.000 | 0.2306 | No | ||
| 45 | PURA | 9670 | 10425 | -0.001 | 0.1951 | No | ||
| 46 | CALD1 | 4273 8463 | 10526 | -0.001 | 0.1898 | No | ||
| 47 | RERG | 16949 | 10685 | -0.002 | 0.1814 | No | ||
| 48 | RARB | 3011 21915 | 10781 | -0.002 | 0.1764 | No | ||
| 49 | GGN | 2472 1171 18312 | 11258 | -0.003 | 0.1510 | No | ||
| 50 | APBA2BP | 14385 | 12387 | -0.007 | 0.0907 | No | ||
| 51 | GPC4 | 24153 | 12456 | -0.007 | 0.0876 | No | ||
| 52 | SH3GL3 | 18201 | 12586 | -0.008 | 0.0812 | No | ||
| 53 | POU4F3 | 9604 | 13266 | -0.012 | 0.0455 | No | ||
| 54 | TIA1 | 5752 | 13326 | -0.012 | 0.0432 | No | ||
| 55 | HOXB8 | 9111 | 13390 | -0.013 | 0.0407 | No | ||
| 56 | HIVEP1 | 8486 | 13756 | -0.016 | 0.0222 | No | ||
| 57 | SP6 | 8178 13678 13679 | 13861 | -0.017 | 0.0179 | No | ||
| 58 | PAPPA | 16190 | 14019 | -0.019 | 0.0108 | No | ||
| 59 | RHOQ | 23142 | 14088 | -0.021 | 0.0087 | No | ||
| 60 | BNC2 | 15850 | 14331 | -0.025 | -0.0026 | No | ||
| 61 | NEUROG1 | 21446 | 14365 | -0.026 | -0.0025 | No | ||
| 62 | JMJD1C | 11531 19996 | 14743 | -0.036 | -0.0201 | No | ||
| 63 | ALCAM | 4367 | 15121 | -0.052 | -0.0367 | No | ||
| 64 | SRPK2 | 5513 | 15728 | -0.101 | -0.0620 | No | ||
| 65 | SLC25A28 | 23669 | 16245 | -0.204 | -0.0750 | No | ||
| 66 | RHOBTB1 | 12789 | 16603 | -0.307 | -0.0718 | No | ||
| 67 | RGS12 | 16872 3534 | 17073 | -0.472 | -0.0627 | No | ||
| 68 | PUM2 | 2071 8161 | 17340 | -0.599 | -0.0334 | No | ||
| 69 | HNRPA0 | 13382 | 17408 | -0.644 | 0.0099 | No | ||
| 70 | HOXB4 | 20686 | 17598 | -0.758 | 0.0549 | No |