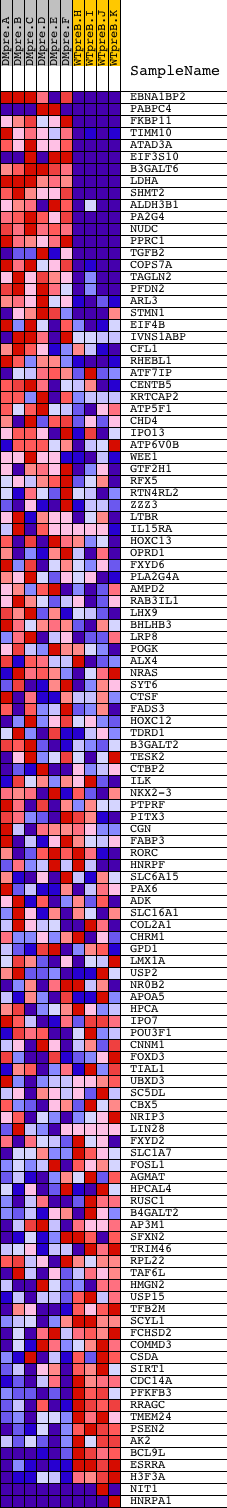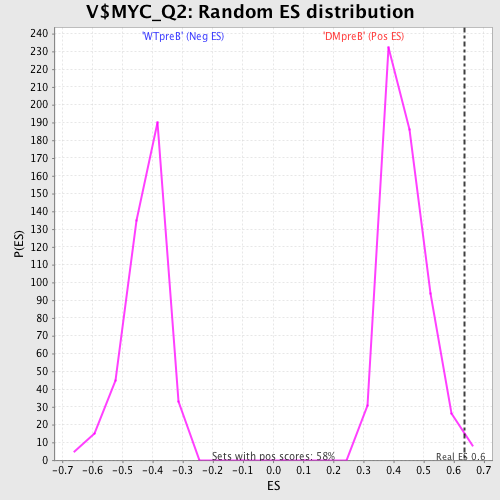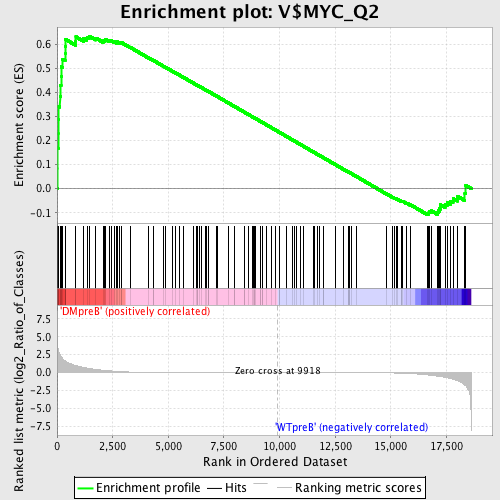
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.6346547 |
| Normalized Enrichment Score (NES) | 1.4520442 |
| Nominal p-value | 0.012131716 |
| FDR q-value | 0.49650297 |
| FWER p-Value | 0.962 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EBNA1BP2 | 8112 7599 12770 | 10 | 4.721 | 0.0855 | Yes | ||
| 2 | PABPC4 | 10510 2418 2548 | 11 | 4.632 | 0.1700 | Yes | ||
| 3 | FKBP11 | 22136 | 46 | 3.327 | 0.2288 | Yes | ||
| 4 | TIMM10 | 11390 2158 | 50 | 3.231 | 0.2876 | Yes | ||
| 5 | ATAD3A | 8440 | 57 | 3.079 | 0.3434 | Yes | ||
| 6 | EIF3S10 | 4659 8887 | 129 | 2.520 | 0.3855 | Yes | ||
| 7 | B3GALT6 | 4391 | 148 | 2.442 | 0.4290 | Yes | ||
| 8 | LDHA | 9269 | 178 | 2.257 | 0.4686 | Yes | ||
| 9 | SHMT2 | 3307 19601 | 188 | 2.155 | 0.5074 | Yes | ||
| 10 | ALDH3B1 | 12569 23949 | 253 | 1.928 | 0.5391 | Yes | ||
| 11 | PA2G4 | 5265 | 360 | 1.642 | 0.5633 | Yes | ||
| 12 | NUDC | 9495 | 368 | 1.624 | 0.5925 | Yes | ||
| 13 | PPRC1 | 23808 | 379 | 1.599 | 0.6212 | Yes | ||
| 14 | TGFB2 | 5743 | 839 | 0.982 | 0.6143 | Yes | ||
| 15 | COPS7A | 11221 16997 | 847 | 0.973 | 0.6316 | Yes | ||
| 16 | TAGLN2 | 14043 | 1182 | 0.722 | 0.6267 | Yes | ||
| 17 | PFDN2 | 9551 | 1343 | 0.621 | 0.6294 | Yes | ||
| 18 | ARL3 | 23652 | 1439 | 0.570 | 0.6347 | Yes | ||
| 19 | STMN1 | 9261 | 1739 | 0.442 | 0.6266 | No | ||
| 20 | EIF4B | 13279 7979 | 2068 | 0.332 | 0.6149 | No | ||
| 21 | IVNS1ABP | 4384 4050 | 2138 | 0.308 | 0.6168 | No | ||
| 22 | CFL1 | 4516 | 2158 | 0.303 | 0.6213 | No | ||
| 23 | RHEBL1 | 12781 | 2336 | 0.255 | 0.6163 | No | ||
| 24 | ATF7IP | 12028 13038 | 2434 | 0.228 | 0.6152 | No | ||
| 25 | CENTB5 | 15961 | 2560 | 0.198 | 0.6121 | No | ||
| 26 | KRTCAP2 | 15541 | 2686 | 0.170 | 0.6084 | No | ||
| 27 | ATP5F1 | 15212 | 2692 | 0.168 | 0.6112 | No | ||
| 28 | CHD4 | 8418 17281 4225 | 2821 | 0.139 | 0.6069 | No | ||
| 29 | IPO13 | 15784 | 2900 | 0.126 | 0.6049 | No | ||
| 30 | ATP6V0B | 15786 | 2902 | 0.126 | 0.6072 | No | ||
| 31 | WEE1 | 18127 | 3278 | 0.080 | 0.5884 | No | ||
| 32 | GTF2H1 | 4069 18236 | 4104 | 0.038 | 0.5445 | No | ||
| 33 | RFX5 | 7018 | 4349 | 0.031 | 0.5318 | No | ||
| 34 | RTN4RL2 | 14544 | 4353 | 0.031 | 0.5322 | No | ||
| 35 | ZZZ3 | 15389 8445 4253 8444 1761 | 4788 | 0.024 | 0.5092 | No | ||
| 36 | LTBR | 16993 | 4892 | 0.022 | 0.5040 | No | ||
| 37 | IL15RA | 2899 2717 15118 2792 | 5170 | 0.019 | 0.4894 | No | ||
| 38 | HOXC13 | 22344 | 5316 | 0.018 | 0.4819 | No | ||
| 39 | OPRD1 | 15737 | 5479 | 0.016 | 0.4734 | No | ||
| 40 | FXYD6 | 19471 | 5664 | 0.015 | 0.4638 | No | ||
| 41 | PLA2G4A | 13809 | 6112 | 0.012 | 0.4398 | No | ||
| 42 | AMPD2 | 1931 15197 | 6243 | 0.011 | 0.4330 | No | ||
| 43 | RAB3IL1 | 3723 13229 | 6292 | 0.011 | 0.4306 | No | ||
| 44 | LHX9 | 9276 | 6330 | 0.011 | 0.4288 | No | ||
| 45 | BHLHB3 | 16935 | 6406 | 0.010 | 0.4249 | No | ||
| 46 | LRP8 | 16149 2425 | 6505 | 0.010 | 0.4198 | No | ||
| 47 | POGK | 7739 | 6677 | 0.009 | 0.4107 | No | ||
| 48 | ALX4 | 4379 | 6715 | 0.009 | 0.4089 | No | ||
| 49 | NRAS | 5191 | 6782 | 0.009 | 0.4055 | No | ||
| 50 | SYT6 | 13437 7042 12040 | 6826 | 0.009 | 0.4033 | No | ||
| 51 | CTSF | 23772 | 7150 | 0.007 | 0.3860 | No | ||
| 52 | FADS3 | 7214 | 7228 | 0.007 | 0.3820 | No | ||
| 53 | HOXC12 | 22343 | 7686 | 0.006 | 0.3574 | No | ||
| 54 | TDRD1 | 13510 23818 | 7958 | 0.005 | 0.3428 | No | ||
| 55 | B3GALT2 | 6498 | 7969 | 0.005 | 0.3423 | No | ||
| 56 | TESK2 | 10504 | 8420 | 0.004 | 0.3181 | No | ||
| 57 | CTBP2 | 17591 1137 | 8428 | 0.003 | 0.3178 | No | ||
| 58 | ILK | 9177 1618 | 8621 | 0.003 | 0.3074 | No | ||
| 59 | NKX2-3 | 5177 23845 | 8761 | 0.003 | 0.3000 | No | ||
| 60 | PTPRF | 5331 2545 | 8807 | 0.003 | 0.2976 | No | ||
| 61 | PITX3 | 9571 | 8867 | 0.002 | 0.2944 | No | ||
| 62 | CGN | 12896 12897 7701 | 8870 | 0.002 | 0.2944 | No | ||
| 63 | FABP3 | 8945 | 8929 | 0.002 | 0.2913 | No | ||
| 64 | RORC | 9732 | 9123 | 0.002 | 0.2809 | No | ||
| 65 | HNRPF | 13607 | 9135 | 0.002 | 0.2803 | No | ||
| 66 | SLC6A15 | 8335 3321 | 9210 | 0.002 | 0.2764 | No | ||
| 67 | PAX6 | 5223 | 9241 | 0.002 | 0.2748 | No | ||
| 68 | ADK | 3302 3454 8555 | 9399 | 0.001 | 0.2663 | No | ||
| 69 | SLC16A1 | 15468 | 9647 | 0.001 | 0.2530 | No | ||
| 70 | COL2A1 | 22145 4542 | 9823 | 0.000 | 0.2435 | No | ||
| 71 | CHRM1 | 23943 | 9991 | -0.000 | 0.2345 | No | ||
| 72 | GPD1 | 22361 | 10004 | -0.000 | 0.2338 | No | ||
| 73 | LMX1A | 14062 3939 | 10327 | -0.001 | 0.2164 | No | ||
| 74 | USP2 | 19480 3052 3043 | 10584 | -0.002 | 0.2026 | No | ||
| 75 | NR0B2 | 16050 | 10663 | -0.002 | 0.1985 | No | ||
| 76 | APOA5 | 19469 | 10770 | -0.002 | 0.1928 | No | ||
| 77 | HPCA | 9118 | 10928 | -0.002 | 0.1843 | No | ||
| 78 | IPO7 | 6130 | 11087 | -0.003 | 0.1758 | No | ||
| 79 | POU3F1 | 16093 | 11539 | -0.004 | 0.1515 | No | ||
| 80 | CNNM1 | 23846 | 11566 | -0.004 | 0.1502 | No | ||
| 81 | FOXD3 | 9088 | 11682 | -0.005 | 0.1441 | No | ||
| 82 | TIAL1 | 996 5753 | 11789 | -0.005 | 0.1384 | No | ||
| 83 | UBXD3 | 15701 | 11966 | -0.005 | 0.1290 | No | ||
| 84 | SC5DL | 6165 | 11980 | -0.006 | 0.1284 | No | ||
| 85 | CBX5 | 22108 4485 | 12504 | -0.008 | 0.1003 | No | ||
| 86 | NRIP3 | 17679 | 12891 | -0.010 | 0.0796 | No | ||
| 87 | LIN28 | 15723 | 13077 | -0.011 | 0.0698 | No | ||
| 88 | FXYD2 | 3038 19470 3068 3039 | 13101 | -0.011 | 0.0687 | No | ||
| 89 | SLC1A7 | 16147 | 13126 | -0.011 | 0.0676 | No | ||
| 90 | FOSL1 | 23779 | 13138 | -0.011 | 0.0672 | No | ||
| 91 | AGMAT | 16002 | 13215 | -0.012 | 0.0633 | No | ||
| 92 | HPCAL4 | 16096 | 13472 | -0.013 | 0.0497 | No | ||
| 93 | RUSC1 | 1853 15283 | 14824 | -0.039 | -0.0226 | No | ||
| 94 | B4GALT2 | 11990 | 15092 | -0.051 | -0.0361 | No | ||
| 95 | AP3M1 | 7059 | 15168 | -0.054 | -0.0392 | No | ||
| 96 | SFXN2 | 3707 8255 3678 13588 | 15251 | -0.059 | -0.0425 | No | ||
| 97 | TRIM46 | 1779 15281 | 15315 | -0.063 | -0.0448 | No | ||
| 98 | RPL22 | 15979 | 15485 | -0.076 | -0.0525 | No | ||
| 99 | TAF6L | 5923 | 15498 | -0.077 | -0.0518 | No | ||
| 100 | HMGN2 | 9095 | 15538 | -0.081 | -0.0524 | No | ||
| 101 | USP15 | 4758 3377 | 15712 | -0.099 | -0.0600 | No | ||
| 102 | TFB2M | 4024 9092 4083 | 15864 | -0.123 | -0.0659 | No | ||
| 103 | SCYL1 | 23986 | 16631 | -0.319 | -0.1015 | No | ||
| 104 | FCHSD2 | 18172 | 16713 | -0.344 | -0.0996 | No | ||
| 105 | COMMD3 | 73 | 16724 | -0.346 | -0.0938 | No | ||
| 106 | CSDA | 1163 16966 1134 | 16817 | -0.376 | -0.0919 | No | ||
| 107 | SIRT1 | 19745 | 17091 | -0.483 | -0.0979 | No | ||
| 108 | CDC14A | 15184 | 17137 | -0.504 | -0.0911 | No | ||
| 109 | PFKFB3 | 2748 | 17179 | -0.520 | -0.0838 | No | ||
| 110 | RRAGC | 12015 | 17217 | -0.532 | -0.0761 | No | ||
| 111 | TMEM24 | 19150 | 17219 | -0.532 | -0.0665 | No | ||
| 112 | PSEN2 | 13736 | 17438 | -0.666 | -0.0661 | No | ||
| 113 | AK2 | 8563 2479 16073 | 17531 | -0.720 | -0.0580 | No | ||
| 114 | BCL9L | 19475 | 17687 | -0.820 | -0.0514 | No | ||
| 115 | ESRRA | 23802 | 17796 | -0.905 | -0.0408 | No | ||
| 116 | H3F3A | 4838 | 17994 | -1.097 | -0.0314 | No | ||
| 117 | NIT1 | 11293 933 | 18330 | -1.725 | -0.0181 | No | ||
| 118 | HNRPA1 | 9103 4860 | 18370 | -1.836 | 0.0133 | No |