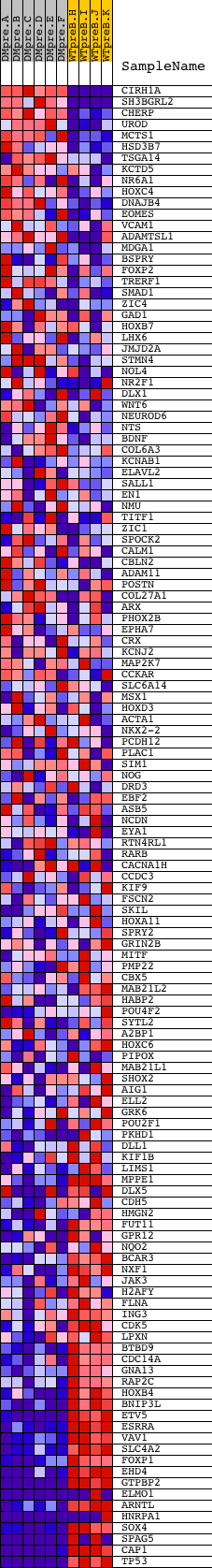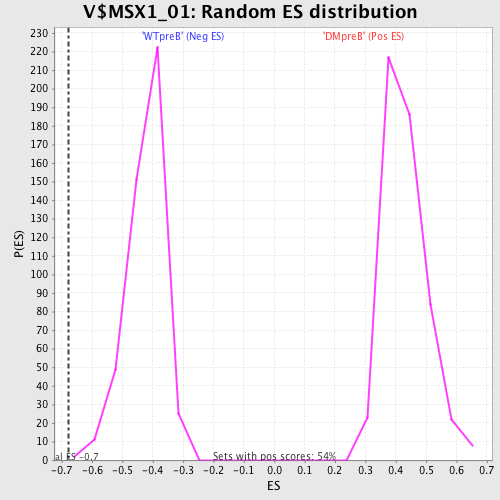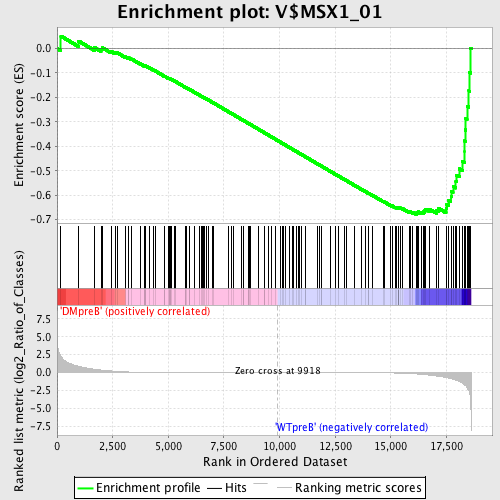
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$MSX1_01 |
| Enrichment Score (ES) | -0.67885715 |
| Normalized Enrichment Score (NES) | -1.6051801 |
| Nominal p-value | 0.002173913 |
| FDR q-value | 0.052014433 |
| FWER p-Value | 0.242 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CIRH1A | 18478 | 172 | 2.288 | 0.0506 | No | ||
| 2 | SH3BGRL2 | 5601 | 967 | 0.866 | 0.0304 | No | ||
| 3 | CHERP | 11367 | 1682 | 0.465 | 0.0039 | No | ||
| 4 | UROD | 15791 | 1974 | 0.360 | -0.0024 | No | ||
| 5 | MCTS1 | 24351 | 2043 | 0.339 | 0.0028 | No | ||
| 6 | HSD3B7 | 4158 | 2422 | 0.231 | -0.0116 | No | ||
| 7 | TSGA14 | 17194 | 2614 | 0.187 | -0.0170 | No | ||
| 8 | KCTD5 | 23096 | 2693 | 0.168 | -0.0168 | No | ||
| 9 | NR6A1 | 14596 | 3057 | 0.101 | -0.0338 | No | ||
| 10 | HOXC4 | 4864 | 3198 | 0.085 | -0.0391 | No | ||
| 11 | DNAJB4 | 12451 1916 1832 | 3224 | 0.084 | -0.0383 | No | ||
| 12 | EOMES | 4669 | 3344 | 0.073 | -0.0428 | No | ||
| 13 | VCAM1 | 5851 | 3731 | 0.051 | -0.0624 | No | ||
| 14 | ADAMTSL1 | 16181 | 3924 | 0.043 | -0.0717 | No | ||
| 15 | MDGA1 | 13231 | 3925 | 0.043 | -0.0705 | No | ||
| 16 | BSPRY | 5307 | 3964 | 0.042 | -0.0715 | No | ||
| 17 | FOXP2 | 4312 17523 8523 | 3965 | 0.042 | -0.0704 | No | ||
| 18 | TRERF1 | 1546 23213 | 4149 | 0.036 | -0.0793 | No | ||
| 19 | SMAD1 | 18831 922 | 4321 | 0.032 | -0.0878 | No | ||
| 20 | ZIC4 | 5992 | 4401 | 0.030 | -0.0912 | No | ||
| 21 | GAD1 | 14993 2690 | 4810 | 0.023 | -0.1127 | No | ||
| 22 | HOXB7 | 666 9110 | 4847 | 0.023 | -0.1141 | No | ||
| 23 | LHX6 | 9275 2914 | 5012 | 0.021 | -0.1224 | No | ||
| 24 | JMJD2A | 10505 6058 | 5046 | 0.021 | -0.1236 | No | ||
| 25 | STMN4 | 21977 | 5057 | 0.020 | -0.1236 | No | ||
| 26 | NOL4 | 2028 11432 | 5092 | 0.020 | -0.1250 | No | ||
| 27 | NR2F1 | 7015 21401 | 5097 | 0.020 | -0.1246 | No | ||
| 28 | DLX1 | 14988 | 5157 | 0.019 | -0.1273 | No | ||
| 29 | WNT6 | 5881 | 5282 | 0.018 | -0.1336 | No | ||
| 30 | NEUROD6 | 17138 | 5322 | 0.018 | -0.1352 | No | ||
| 31 | NTS | 19635 3297 | 5751 | 0.014 | -0.1580 | No | ||
| 32 | BDNF | 14926 2797 | 5772 | 0.014 | -0.1587 | No | ||
| 33 | COL6A3 | 4546 13885 13884 | 5816 | 0.013 | -0.1607 | No | ||
| 34 | KCNAB1 | 1791 15577 | 5930 | 0.013 | -0.1665 | No | ||
| 35 | ELAVL2 | 15839 | 5949 | 0.013 | -0.1671 | No | ||
| 36 | SALL1 | 18796 12207 | 6196 | 0.011 | -0.1801 | No | ||
| 37 | EN1 | 387 14155 | 6409 | 0.010 | -0.1913 | No | ||
| 38 | NMU | 16506 | 6503 | 0.010 | -0.1961 | No | ||
| 39 | TITF1 | 10180 21062 | 6524 | 0.010 | -0.1969 | No | ||
| 40 | ZIC1 | 19040 | 6577 | 0.010 | -0.1995 | No | ||
| 41 | SPOCK2 | 13585 | 6642 | 0.009 | -0.2027 | No | ||
| 42 | CALM1 | 21184 | 6700 | 0.009 | -0.2055 | No | ||
| 43 | CBLN2 | 23498 | 6788 | 0.009 | -0.2100 | No | ||
| 44 | ADAM11 | 4340 1432 | 6991 | 0.008 | -0.2207 | No | ||
| 45 | POSTN | 1801 11927 | 7049 | 0.008 | -0.2236 | No | ||
| 46 | COL27A1 | 6885 | 7714 | 0.005 | -0.2594 | No | ||
| 47 | ARX | 24290 | 7842 | 0.005 | -0.2661 | No | ||
| 48 | PHOX2B | 16524 | 7926 | 0.005 | -0.2705 | No | ||
| 49 | EPHA7 | 8909 4674 | 8308 | 0.004 | -0.2910 | No | ||
| 50 | CRX | 17961 4694 | 8361 | 0.004 | -0.2937 | No | ||
| 51 | KCNJ2 | 20612 | 8380 | 0.004 | -0.2946 | No | ||
| 52 | MAP2K7 | 6453 | 8614 | 0.003 | -0.3071 | No | ||
| 53 | CCKAR | 16531 | 8624 | 0.003 | -0.3075 | No | ||
| 54 | SLC6A14 | 12168 | 8653 | 0.003 | -0.3090 | No | ||
| 55 | MSX1 | 16547 | 8673 | 0.003 | -0.3099 | No | ||
| 56 | HOXD3 | 9115 | 9064 | 0.002 | -0.3310 | No | ||
| 57 | ACTA1 | 18715 | 9301 | 0.001 | -0.3437 | No | ||
| 58 | NKX2-2 | 5176 | 9308 | 0.001 | -0.3440 | No | ||
| 59 | PCDH12 | 7005 | 9487 | 0.001 | -0.3536 | No | ||
| 60 | PLAC1 | 12079 | 9642 | 0.001 | -0.3619 | No | ||
| 61 | SIM1 | 20031 | 9814 | 0.000 | -0.3712 | No | ||
| 62 | NOG | 13663 | 10030 | -0.000 | -0.3828 | No | ||
| 63 | DRD3 | 22750 | 10108 | -0.000 | -0.3869 | No | ||
| 64 | EBF2 | 21975 | 10186 | -0.001 | -0.3911 | No | ||
| 65 | ASB5 | 13322 | 10274 | -0.001 | -0.3958 | No | ||
| 66 | NCDN | 15756 2487 6471 | 10452 | -0.001 | -0.4053 | No | ||
| 67 | EYA1 | 4695 4061 | 10600 | -0.002 | -0.4132 | No | ||
| 68 | RTN4RL1 | 20782 | 10603 | -0.002 | -0.4133 | No | ||
| 69 | RARB | 3011 21915 | 10781 | -0.002 | -0.4228 | No | ||
| 70 | CACNA1H | 23077 | 10856 | -0.002 | -0.4268 | No | ||
| 71 | CCDC3 | 15122 | 10903 | -0.002 | -0.4292 | No | ||
| 72 | KIF9 | 19292 | 10999 | -0.003 | -0.4343 | No | ||
| 73 | FSCN2 | 20569 | 11163 | -0.003 | -0.4430 | No | ||
| 74 | SKIL | 15617 | 11169 | -0.003 | -0.4432 | No | ||
| 75 | HOXA11 | 17146 | 11691 | -0.005 | -0.4712 | No | ||
| 76 | SPRY2 | 21725 | 11810 | -0.005 | -0.4775 | No | ||
| 77 | GRIN2B | 16957 | 11902 | -0.005 | -0.4823 | No | ||
| 78 | MITF | 17349 | 11903 | -0.005 | -0.4821 | No | ||
| 79 | PMP22 | 9594 1312 | 12282 | -0.007 | -0.5024 | No | ||
| 80 | CBX5 | 22108 4485 | 12504 | -0.008 | -0.5142 | No | ||
| 81 | MAB21L2 | 15305 | 12656 | -0.009 | -0.5221 | No | ||
| 82 | HABP2 | 10364 23825 | 12911 | -0.010 | -0.5356 | No | ||
| 83 | POU4F2 | 9603 5276 | 13020 | -0.010 | -0.5412 | No | ||
| 84 | SYTL2 | 18190 1539 3730 | 13371 | -0.013 | -0.5598 | No | ||
| 85 | A2BP1 | 11212 1652 1689 | 13682 | -0.015 | -0.5762 | No | ||
| 86 | HOXC6 | 22339 2302 | 13871 | -0.017 | -0.5859 | No | ||
| 87 | PIPOX | 20340 | 13990 | -0.019 | -0.5918 | No | ||
| 88 | MAB21L1 | 15589 5046 | 13994 | -0.019 | -0.5914 | No | ||
| 89 | SHOX2 | 1802 5432 | 14158 | -0.022 | -0.5997 | No | ||
| 90 | AIG1 | 7272 | 14670 | -0.034 | -0.6264 | No | ||
| 91 | ELL2 | 5329 | 14712 | -0.035 | -0.6277 | No | ||
| 92 | GRK6 | 6449 | 14996 | -0.046 | -0.6418 | No | ||
| 93 | POU2F1 | 5275 3989 4065 4010 | 15053 | -0.048 | -0.6436 | No | ||
| 94 | PKHD1 | 6292 4106 | 15216 | -0.057 | -0.6509 | No | ||
| 95 | DLL1 | 23119 | 15244 | -0.059 | -0.6508 | No | ||
| 96 | KIF1B | 15671 2476 4950 15673 | 15329 | -0.064 | -0.6537 | No | ||
| 97 | LIMS1 | 8493 4290 | 15332 | -0.064 | -0.6521 | No | ||
| 98 | MPPE1 | 23417 2013 | 15333 | -0.064 | -0.6504 | No | ||
| 99 | DLX5 | 17219 1058 | 15351 | -0.065 | -0.6496 | No | ||
| 100 | CDH5 | 18512 | 15432 | -0.071 | -0.6521 | No | ||
| 101 | HMGN2 | 9095 | 15538 | -0.081 | -0.6557 | No | ||
| 102 | FUT11 | 13084 | 15848 | -0.120 | -0.6692 | No | ||
| 103 | GPR12 | 16291 | 15875 | -0.125 | -0.6674 | No | ||
| 104 | NQO2 | 9469 3167 | 15995 | -0.146 | -0.6700 | No | ||
| 105 | BCAR3 | 15432 | 16160 | -0.183 | -0.6741 | Yes | ||
| 106 | NXF1 | 11986 | 16194 | -0.190 | -0.6709 | Yes | ||
| 107 | JAK3 | 9198 4936 | 16236 | -0.199 | -0.6679 | Yes | ||
| 108 | H2AFY | 21447 | 16389 | -0.241 | -0.6698 | Yes | ||
| 109 | FLNA | 24129 | 16483 | -0.269 | -0.6678 | Yes | ||
| 110 | ING3 | 17514 1019 | 16529 | -0.284 | -0.6628 | Yes | ||
| 111 | CDK5 | 16591 | 16566 | -0.296 | -0.6569 | Yes | ||
| 112 | LPXN | 8399 | 16734 | -0.351 | -0.6568 | Yes | ||
| 113 | BTBD9 | 23040 | 17046 | -0.463 | -0.6615 | Yes | ||
| 114 | CDC14A | 15184 | 17137 | -0.504 | -0.6531 | Yes | ||
| 115 | GNA13 | 20617 | 17482 | -0.692 | -0.6536 | Yes | ||
| 116 | RAP2C | 24156 | 17516 | -0.713 | -0.6367 | Yes | ||
| 117 | HOXB4 | 20686 | 17598 | -0.758 | -0.6212 | Yes | ||
| 118 | BNIP3L | 21775 | 17704 | -0.839 | -0.6049 | Yes | ||
| 119 | ETV5 | 22630 | 17725 | -0.855 | -0.5836 | Yes | ||
| 120 | ESRRA | 23802 | 17796 | -0.905 | -0.5637 | Yes | ||
| 121 | VAV1 | 23173 | 17925 | -1.033 | -0.5435 | Yes | ||
| 122 | SLC4A2 | 9828 | 17961 | -1.069 | -0.5174 | Yes | ||
| 123 | FOXP1 | 4242 | 18090 | -1.220 | -0.4924 | Yes | ||
| 124 | EHD4 | 8275 | 18216 | -1.417 | -0.4620 | Yes | ||
| 125 | GTPBP2 | 23221 | 18322 | -1.708 | -0.4229 | Yes | ||
| 126 | ELMO1 | 3275 8939 3202 3271 3254 3235 3164 3175 3274 3188 3293 3214 3218 3239 | 18325 | -1.717 | -0.3780 | Yes | ||
| 127 | ARNTL | 18119 | 18345 | -1.760 | -0.3329 | Yes | ||
| 128 | HNRPA1 | 9103 4860 | 18370 | -1.836 | -0.2861 | Yes | ||
| 129 | SOX4 | 5479 | 18427 | -2.035 | -0.2358 | Yes | ||
| 130 | SPAG5 | 20760 1341 | 18508 | -2.540 | -0.1736 | Yes | ||
| 131 | CAP1 | 8684 | 18546 | -2.934 | -0.0987 | Yes | ||
| 132 | TP53 | 20822 | 18582 | -3.909 | 0.0018 | Yes |