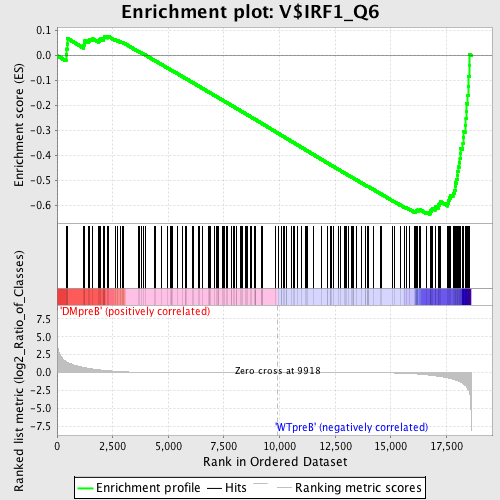
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$IRF1_Q6 |
| Enrichment Score (ES) | -0.6352204 |
| Normalized Enrichment Score (NES) | -1.5605855 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.075747915 |
| FWER p-Value | 0.428 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CNOT4 | 7010 1022 | 403 | 1.537 | 0.0026 | No | ||
| 2 | LY86 | 21665 | 416 | 1.514 | 0.0261 | No | ||
| 3 | PSME2 | 5301 | 465 | 1.425 | 0.0462 | No | ||
| 4 | ZMPSTE24 | 6061 6060 10509 15769 | 475 | 1.407 | 0.0681 | No | ||
| 5 | RPS19 | 5398 | 1201 | 0.713 | 0.0402 | No | ||
| 6 | VAMP8 | 1034 17117 | 1215 | 0.705 | 0.0507 | No | ||
| 7 | TRIP13 | 21406 | 1248 | 0.680 | 0.0598 | No | ||
| 8 | ITGB7 | 22112 | 1427 | 0.577 | 0.0594 | No | ||
| 9 | RAD1 | 22507 | 1469 | 0.555 | 0.0660 | No | ||
| 10 | ATP2C1 | 10660 6178 | 1606 | 0.497 | 0.0665 | No | ||
| 11 | FNTB | 21235 | 1853 | 0.399 | 0.0595 | No | ||
| 12 | RPA2 | 2330 16057 | 1910 | 0.383 | 0.0626 | No | ||
| 13 | SKP1A | 20889 | 1931 | 0.374 | 0.0675 | No | ||
| 14 | KIF3A | 9219 4953 | 2082 | 0.326 | 0.0645 | No | ||
| 15 | PSMB3 | 11180 | 2094 | 0.322 | 0.0691 | No | ||
| 16 | EPN2 | 20413 945 946 955 | 2108 | 0.316 | 0.0734 | No | ||
| 17 | GFI1 | 16451 | 2124 | 0.311 | 0.0775 | No | ||
| 18 | VDAC2 | 22081 | 2265 | 0.270 | 0.0743 | No | ||
| 19 | GPR65 | 21187 | 2292 | 0.265 | 0.0771 | No | ||
| 20 | PROX1 | 9623 | 2617 | 0.186 | 0.0624 | No | ||
| 21 | NT5C3 | 17136 | 2720 | 0.162 | 0.0595 | No | ||
| 22 | LAMC1 | 13805 | 2846 | 0.135 | 0.0549 | No | ||
| 23 | ADAM15 | 15275 | 2924 | 0.123 | 0.0527 | No | ||
| 24 | CUL2 | 23630 | 3002 | 0.110 | 0.0502 | No | ||
| 25 | ZRANB1 | 6884 11702 | 3635 | 0.056 | 0.0168 | No | ||
| 26 | SLC26A6 | 19304 | 3702 | 0.052 | 0.0141 | No | ||
| 27 | PPP1R12C | 6115 17987 | 3788 | 0.049 | 0.0103 | No | ||
| 28 | PKIG | 9577 655 | 3896 | 0.044 | 0.0052 | No | ||
| 29 | SNAI1 | 14726 | 3955 | 0.042 | 0.0027 | No | ||
| 30 | CALU | 1124 1149 8683 1162 4472 | 4373 | 0.031 | -0.0194 | No | ||
| 31 | FES | 17779 3949 | 4419 | 0.030 | -0.0214 | No | ||
| 32 | HOXA10 | 17145 989 | 4684 | 0.025 | -0.0353 | No | ||
| 33 | NCAM2 | 9445 5150 | 4939 | 0.022 | -0.0488 | No | ||
| 34 | HOXA13 | 9107 | 5078 | 0.020 | -0.0559 | No | ||
| 35 | CDX1 | 23429 | 5131 | 0.020 | -0.0584 | No | ||
| 36 | DLX1 | 14988 | 5157 | 0.019 | -0.0595 | No | ||
| 37 | ERBB2 | 8913 | 5205 | 0.019 | -0.0617 | No | ||
| 38 | TNFSF13B | 6291 10792 | 5409 | 0.017 | -0.0725 | No | ||
| 39 | USF1 | 5832 10260 10261 | 5657 | 0.015 | -0.0856 | No | ||
| 40 | ZNF385 | 22106 2297 | 5757 | 0.014 | -0.0908 | No | ||
| 41 | ZMYND17 | 21909 | 5783 | 0.014 | -0.0919 | No | ||
| 42 | TCF15 | 14798 | 5812 | 0.013 | -0.0932 | No | ||
| 43 | SERPINI1 | 340 | 6094 | 0.012 | -0.1083 | No | ||
| 44 | NR3C2 | 18562 13798 | 6133 | 0.012 | -0.1102 | No | ||
| 45 | CBARA1 | 20015 | 6367 | 0.010 | -0.1226 | No | ||
| 46 | IL18BP | 17730 | 6407 | 0.010 | -0.1246 | No | ||
| 47 | TITF1 | 10180 21062 | 6524 | 0.010 | -0.1307 | No | ||
| 48 | NRAS | 5191 | 6782 | 0.009 | -0.1445 | No | ||
| 49 | HNRPA2B1 | 1187 7001 7000 1040 | 6856 | 0.008 | -0.1484 | No | ||
| 50 | OTX1 | 20517 | 6868 | 0.008 | -0.1488 | No | ||
| 51 | MAMDC1 | 21054 6798 | 6882 | 0.008 | -0.1494 | No | ||
| 52 | DSPP | 8867 | 7068 | 0.008 | -0.1593 | No | ||
| 53 | GLRA1 | 9021 | 7165 | 0.007 | -0.1644 | No | ||
| 54 | MYL3 | 19291 | 7219 | 0.007 | -0.1672 | No | ||
| 55 | DPP10 | 13850 | 7232 | 0.007 | -0.1677 | No | ||
| 56 | IL27 | 17636 | 7244 | 0.007 | -0.1682 | No | ||
| 57 | OTP | 21582 | 7427 | 0.006 | -0.1779 | No | ||
| 58 | TRIM23 | 8165 3230 | 7457 | 0.006 | -0.1794 | No | ||
| 59 | STX16 | 10464 | 7540 | 0.006 | -0.1838 | No | ||
| 60 | QKI | 23128 5340 | 7625 | 0.006 | -0.1882 | No | ||
| 61 | STAG1 | 5520 9905 2987 | 7662 | 0.006 | -0.1901 | No | ||
| 62 | FGB | 15309 | 7816 | 0.005 | -0.1983 | No | ||
| 63 | ELMO3 | 10629 18495 | 7850 | 0.005 | -0.2000 | No | ||
| 64 | PHOX2B | 16524 | 7926 | 0.005 | -0.2040 | No | ||
| 65 | B3GALT2 | 6498 | 7969 | 0.005 | -0.2062 | No | ||
| 66 | CBX3 | 8705 4484 | 8060 | 0.004 | -0.2110 | No | ||
| 67 | HOXD13 | 9114 | 8065 | 0.004 | -0.2112 | No | ||
| 68 | FIGN | 14575 | 8244 | 0.004 | -0.2208 | No | ||
| 69 | ELAVL4 | 15805 4889 9137 | 8277 | 0.004 | -0.2225 | No | ||
| 70 | ARHGAP5 | 4412 8625 | 8351 | 0.004 | -0.2264 | No | ||
| 71 | TBX6 | 1993 18075 | 8479 | 0.003 | -0.2332 | No | ||
| 72 | SNTB2 | 3887 18476 | 8521 | 0.003 | -0.2354 | No | ||
| 73 | CES2 | 18505 | 8541 | 0.003 | -0.2363 | No | ||
| 74 | MSX1 | 16547 | 8673 | 0.003 | -0.2434 | No | ||
| 75 | SOX11 | 5477 | 8734 | 0.003 | -0.2466 | No | ||
| 76 | PITX3 | 9571 | 8867 | 0.002 | -0.2537 | No | ||
| 77 | BNIPL | 1890 15249 | 8895 | 0.002 | -0.2552 | No | ||
| 78 | EMX2 | 23806 | 8921 | 0.002 | -0.2565 | No | ||
| 79 | BMP5 | 19376 | 8937 | 0.002 | -0.2573 | No | ||
| 80 | NRXN3 | 2153 5196 | 9167 | 0.002 | -0.2696 | No | ||
| 81 | FGF12 | 1723 22621 | 9207 | 0.002 | -0.2717 | No | ||
| 82 | SEMA5B | 9801 22777 | 9231 | 0.002 | -0.2730 | No | ||
| 83 | PAX6 | 5223 | 9241 | 0.002 | -0.2734 | No | ||
| 84 | SIM1 | 20031 | 9814 | 0.000 | -0.3044 | No | ||
| 85 | FEV | 11153 6434 | 9947 | -0.000 | -0.3116 | No | ||
| 86 | GSH1 | 16622 | 10075 | -0.000 | -0.3185 | No | ||
| 87 | NETO1 | 6372 1970 | 10095 | -0.000 | -0.3195 | No | ||
| 88 | PCDHB1 | 23590 | 10179 | -0.001 | -0.3240 | No | ||
| 89 | LOX | 23438 | 10224 | -0.001 | -0.3264 | No | ||
| 90 | TBX1 | 10039 5630 | 10310 | -0.001 | -0.3310 | No | ||
| 91 | LMX1A | 14062 3939 | 10327 | -0.001 | -0.3318 | No | ||
| 92 | CALD1 | 4273 8463 | 10526 | -0.001 | -0.3425 | No | ||
| 93 | ADORA3 | 4352 | 10635 | -0.002 | -0.3484 | No | ||
| 94 | HGF | 16916 | 10648 | -0.002 | -0.3490 | No | ||
| 95 | GUCY2C | 1140 16954 | 10819 | -0.002 | -0.3582 | No | ||
| 96 | PITX2 | 15424 1878 | 10992 | -0.003 | -0.3675 | No | ||
| 97 | COL9A1 | 3950 4549 | 11154 | -0.003 | -0.3762 | No | ||
| 98 | THRB | 10173 | 11209 | -0.003 | -0.3790 | No | ||
| 99 | PLXNC1 | 7056 | 11247 | -0.003 | -0.3810 | No | ||
| 100 | MMP13 | 19573 3007 | 11510 | -0.004 | -0.3951 | No | ||
| 101 | MITF | 17349 | 11903 | -0.005 | -0.4163 | No | ||
| 102 | UBD | 23239 | 12135 | -0.006 | -0.4288 | No | ||
| 103 | CDH6 | 22320 | 12139 | -0.006 | -0.4288 | No | ||
| 104 | DAPP1 | 11159 6445 6446 | 12268 | -0.007 | -0.4357 | No | ||
| 105 | NHLH2 | 9462 | 12308 | -0.007 | -0.4377 | No | ||
| 106 | PRICKLE2 | 10837 | 12319 | -0.007 | -0.4381 | No | ||
| 107 | ESRRG | 6447 | 12405 | -0.007 | -0.4426 | No | ||
| 108 | MAB21L2 | 15305 | 12656 | -0.009 | -0.4560 | No | ||
| 109 | PCDH9 | 21736 | 12720 | -0.009 | -0.4593 | No | ||
| 110 | GFRA3 | 23469 | 12903 | -0.010 | -0.4690 | No | ||
| 111 | WNT5A | 22066 5880 | 12960 | -0.010 | -0.4719 | No | ||
| 112 | TYR | 17763 | 13005 | -0.010 | -0.4741 | No | ||
| 113 | LIN28 | 15723 | 13077 | -0.011 | -0.4778 | No | ||
| 114 | ASPA | 20354 | 13249 | -0.012 | -0.4869 | No | ||
| 115 | GRID2 | 4803 | 13260 | -0.012 | -0.4872 | No | ||
| 116 | RET | 17028 | 13311 | -0.012 | -0.4898 | No | ||
| 117 | TIA1 | 5752 | 13326 | -0.012 | -0.4903 | No | ||
| 118 | ADAM23 | 10699 4057 | 13463 | -0.013 | -0.4975 | No | ||
| 119 | TMPRSS5 | 19463 | 13681 | -0.015 | -0.5090 | No | ||
| 120 | SIPA1L1 | 5729 | 13867 | -0.017 | -0.5188 | No | ||
| 121 | HTR3B | 19129 3145 | 13872 | -0.017 | -0.5187 | No | ||
| 122 | NFAT5 | 3921 7037 12036 | 13952 | -0.019 | -0.5227 | No | ||
| 123 | SLC12A7 | 21603 | 13973 | -0.019 | -0.5235 | No | ||
| 124 | RCOR1 | 23998 | 13987 | -0.019 | -0.5239 | No | ||
| 125 | MAB21L1 | 15589 5046 | 13994 | -0.019 | -0.5239 | No | ||
| 126 | ENPP6 | 6816 11583 | 14237 | -0.023 | -0.5367 | No | ||
| 127 | JARID2 | 9200 | 14540 | -0.030 | -0.5526 | No | ||
| 128 | DGKA | 3359 19589 | 14560 | -0.030 | -0.5531 | No | ||
| 129 | PTMA | 9657 | 15085 | -0.050 | -0.5808 | No | ||
| 130 | HPCAL1 | 21307 2123 | 15150 | -0.053 | -0.5834 | No | ||
| 131 | TCIRG1 | 3700 3752 23951 | 15423 | -0.070 | -0.5970 | No | ||
| 132 | UGCGL1 | 6721 | 15629 | -0.089 | -0.6067 | No | ||
| 133 | CENTD2 | 1758 18169 2284 | 15682 | -0.095 | -0.6080 | No | ||
| 134 | PPARGC1A | 16533 | 15834 | -0.119 | -0.6143 | No | ||
| 135 | ZADH2 | 23501 | 16067 | -0.161 | -0.6243 | No | ||
| 136 | VIL2 | 5856 | 16130 | -0.174 | -0.6249 | No | ||
| 137 | NEDD4 | 19384 3101 | 16132 | -0.175 | -0.6222 | No | ||
| 138 | MEF2C | 3204 9378 | 16157 | -0.182 | -0.6206 | No | ||
| 139 | TYROBP | 18302 | 16190 | -0.189 | -0.6193 | No | ||
| 140 | TAPBP | 5625 | 16208 | -0.193 | -0.6172 | No | ||
| 141 | TUBD1 | 20718 1399 | 16286 | -0.212 | -0.6180 | No | ||
| 142 | EPS8 | 16948 | 16321 | -0.220 | -0.6163 | No | ||
| 143 | RAC1 | 16302 | 16592 | -0.304 | -0.6261 | No | ||
| 144 | RNF31 | 11206 | 16761 | -0.360 | -0.6295 | Yes | ||
| 145 | PDCD10 | 15314 | 16774 | -0.365 | -0.6243 | Yes | ||
| 146 | P4HA1 | 20017 | 16775 | -0.365 | -0.6185 | Yes | ||
| 147 | DYRK1A | 4649 | 16845 | -0.384 | -0.6161 | Yes | ||
| 148 | CTSS | 15505 | 16861 | -0.390 | -0.6107 | Yes | ||
| 149 | KYNU | 15016 | 16988 | -0.440 | -0.6105 | Yes | ||
| 150 | PSMB10 | 5299 18761 | 16997 | -0.443 | -0.6039 | Yes | ||
| 151 | TTC17 | 13219 | 17125 | -0.500 | -0.6028 | Yes | ||
| 152 | CCR7 | 20254 | 17126 | -0.500 | -0.5949 | Yes | ||
| 153 | NLK | 5179 5178 | 17203 | -0.527 | -0.5906 | Yes | ||
| 154 | RRAGC | 12015 | 17217 | -0.532 | -0.5828 | Yes | ||
| 155 | EHD1 | 23791 | 17561 | -0.738 | -0.5897 | Yes | ||
| 156 | HOXB4 | 20686 | 17598 | -0.758 | -0.5795 | Yes | ||
| 157 | PAIP2 | 12593 | 17632 | -0.779 | -0.5689 | Yes | ||
| 158 | ATP6V0D1 | 3895 8639 3920 | 17671 | -0.807 | -0.5581 | Yes | ||
| 159 | PIAS1 | 7126 | 17797 | -0.905 | -0.5505 | Yes | ||
| 160 | PPM1D | 20721 | 17849 | -0.958 | -0.5380 | Yes | ||
| 161 | MBNL1 | 1921 15582 | 17894 | -1.001 | -0.5244 | Yes | ||
| 162 | MAP3K11 | 11163 | 17905 | -1.013 | -0.5088 | Yes | ||
| 163 | SLC4A2 | 9828 | 17961 | -1.069 | -0.4947 | Yes | ||
| 164 | LSP1 | 18003 | 17990 | -1.095 | -0.4788 | Yes | ||
| 165 | BANK1 | 15159 | 18006 | -1.117 | -0.4618 | Yes | ||
| 166 | FCGR2B | 8959 | 18041 | -1.163 | -0.4451 | Yes | ||
| 167 | TBCC | 23215 | 18068 | -1.197 | -0.4275 | Yes | ||
| 168 | PRKCB1 | 1693 9574 | 18108 | -1.252 | -0.4096 | Yes | ||
| 169 | RIOK3 | 7354 12425 1973 | 18134 | -1.287 | -0.3905 | Yes | ||
| 170 | VGLL4 | 10558 | 18141 | -1.297 | -0.3701 | Yes | ||
| 171 | BMF | 5074 | 18240 | -1.486 | -0.3518 | Yes | ||
| 172 | LMO4 | 15151 | 18255 | -1.527 | -0.3282 | Yes | ||
| 173 | FLI1 | 4729 | 18275 | -1.571 | -0.3042 | Yes | ||
| 174 | RPL23 | 7237 12267 20267 | 18343 | -1.757 | -0.2798 | Yes | ||
| 175 | EIF4A2 | 4660 1679 1645 | 18377 | -1.855 | -0.2520 | Yes | ||
| 176 | WASF2 | 6326 | 18391 | -1.916 | -0.2222 | Yes | ||
| 177 | CD37 | 17835 | 18397 | -1.939 | -0.1916 | Yes | ||
| 178 | MS4A1 | 59 | 18451 | -2.185 | -0.1596 | Yes | ||
| 179 | CNIH | 4536 23971 | 18489 | -2.453 | -0.1225 | Yes | ||
| 180 | CD53 | 4500 | 18501 | -2.502 | -0.0832 | Yes | ||
| 181 | SGK | 9809 3419 | 18533 | -2.771 | -0.0407 | Yes | ||
| 182 | TNFRSF19 | 21799 3033 | 18538 | -2.835 | 0.0042 | Yes |