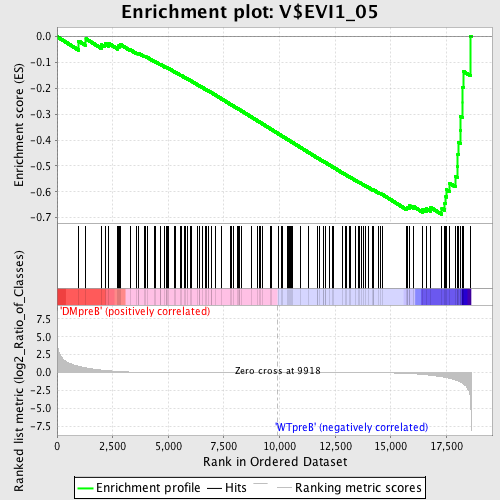
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | V$EVI1_05 |
| Enrichment Score (ES) | -0.68796706 |
| Normalized Enrichment Score (NES) | -1.6381427 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.046795126 |
| FWER p-Value | 0.129 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SH3BGRL2 | 5601 | 967 | 0.866 | -0.0169 | No | ||
| 2 | ISL1 | 21338 | 1297 | 0.646 | -0.0083 | No | ||
| 3 | CHCHD7 | 16280 | 1996 | 0.354 | -0.0316 | No | ||
| 4 | ZFPM2 | 22483 | 2152 | 0.304 | -0.0276 | No | ||
| 5 | WBP5 | 10297 | 2322 | 0.257 | -0.0262 | No | ||
| 6 | SUFU | 6283 | 2730 | 0.159 | -0.0418 | No | ||
| 7 | NDUFA1 | 24170 | 2741 | 0.156 | -0.0359 | No | ||
| 8 | STOML2 | 15902 | 2824 | 0.139 | -0.0347 | No | ||
| 9 | SFRS2 | 9807 20136 | 2845 | 0.136 | -0.0302 | No | ||
| 10 | MAP2K5 | 19088 | 3284 | 0.079 | -0.0507 | No | ||
| 11 | RRAGA | 16180 | 3589 | 0.058 | -0.0647 | No | ||
| 12 | ZRANB1 | 6884 11702 | 3635 | 0.056 | -0.0649 | No | ||
| 13 | VAMP1 | 5846 1043 | 3653 | 0.055 | -0.0636 | No | ||
| 14 | MYCT1 | 7572 | 3921 | 0.043 | -0.0763 | No | ||
| 15 | FOXP2 | 4312 17523 8523 | 3965 | 0.042 | -0.0769 | No | ||
| 16 | ORC4L | 11172 6460 | 4060 | 0.039 | -0.0804 | No | ||
| 17 | OLIG3 | 20084 | 4358 | 0.031 | -0.0952 | No | ||
| 18 | GPR22 | 21087 | 4438 | 0.029 | -0.0982 | No | ||
| 19 | FHL3 | 16092 2457 | 4625 | 0.026 | -0.1072 | No | ||
| 20 | XCR1 | 18953 | 4651 | 0.026 | -0.1075 | No | ||
| 21 | HOXB7 | 666 9110 | 4847 | 0.023 | -0.1171 | No | ||
| 22 | IL4 | 9174 | 4896 | 0.022 | -0.1188 | No | ||
| 23 | HOXB3 | 20685 | 4920 | 0.022 | -0.1192 | No | ||
| 24 | NCAM2 | 9445 5150 | 4939 | 0.022 | -0.1193 | No | ||
| 25 | IRX5 | 18527 | 5006 | 0.021 | -0.1220 | No | ||
| 26 | TCF2 | 1395 20727 | 5297 | 0.018 | -0.1369 | No | ||
| 27 | NEUROD6 | 17138 | 5322 | 0.018 | -0.1375 | No | ||
| 28 | PKHD1L1 | 22481 9650 | 5547 | 0.016 | -0.1490 | No | ||
| 29 | SPINK4 | 16241 | 5579 | 0.015 | -0.1500 | No | ||
| 30 | CACNA1D | 4464 8675 | 5734 | 0.014 | -0.1578 | No | ||
| 31 | MID1 | 5097 5098 | 5769 | 0.014 | -0.1591 | No | ||
| 32 | BMP10 | 17376 | 5871 | 0.013 | -0.1640 | No | ||
| 33 | HOXB5 | 20687 | 5996 | 0.012 | -0.1702 | No | ||
| 34 | CREB5 | 10551 | 6058 | 0.012 | -0.1730 | No | ||
| 35 | HOXA1 | 1005 17152 | 6296 | 0.011 | -0.1854 | No | ||
| 36 | GRM3 | 16929 | 6384 | 0.010 | -0.1896 | No | ||
| 37 | IGF1 | 3352 9156 3409 | 6416 | 0.010 | -0.1909 | No | ||
| 38 | TYRO3 | 5811 | 6553 | 0.010 | -0.1979 | No | ||
| 39 | HOXD10 | 14983 | 6660 | 0.009 | -0.2032 | No | ||
| 40 | NEK6 | 15018 | 6722 | 0.009 | -0.2061 | No | ||
| 41 | NRAS | 5191 | 6782 | 0.009 | -0.2090 | No | ||
| 42 | GNAO1 | 4785 3829 | 6928 | 0.008 | -0.2165 | No | ||
| 43 | NTN4 | 19903 | 6936 | 0.008 | -0.2165 | No | ||
| 44 | PFTK1 | 16924 | 7119 | 0.007 | -0.2261 | No | ||
| 45 | HIP2 | 3626 3485 16838 | 7406 | 0.006 | -0.2413 | No | ||
| 46 | ELOVL3 | 23809 | 7812 | 0.005 | -0.2630 | No | ||
| 47 | DNAJC1 | 8854 | 7821 | 0.005 | -0.2632 | No | ||
| 48 | HCN1 | 21557 | 7934 | 0.005 | -0.2690 | No | ||
| 49 | WNT9A | 5709 | 8122 | 0.004 | -0.2790 | No | ||
| 50 | RFX4 | 7721 3410 | 8137 | 0.004 | -0.2796 | No | ||
| 51 | CYP26B1 | 17093 | 8192 | 0.004 | -0.2823 | No | ||
| 52 | ADPRHL1 | 18930 3863 | 8196 | 0.004 | -0.2823 | No | ||
| 53 | ELAVL4 | 15805 4889 9137 | 8277 | 0.004 | -0.2865 | No | ||
| 54 | SOX11 | 5477 | 8734 | 0.003 | -0.3110 | No | ||
| 55 | HOXA3 | 9108 1075 | 9001 | 0.002 | -0.3253 | No | ||
| 56 | SLC37A4 | 8994 | 9108 | 0.002 | -0.3310 | No | ||
| 57 | GPR23 | 13428 | 9158 | 0.002 | -0.3335 | No | ||
| 58 | GPR44 | 23926 | 9220 | 0.002 | -0.3368 | No | ||
| 59 | PI15 | 13586 | 9569 | 0.001 | -0.3555 | No | ||
| 60 | PLAC1 | 12079 | 9642 | 0.001 | -0.3594 | No | ||
| 61 | TSC1 | 7235 | 9972 | -0.000 | -0.3772 | No | ||
| 62 | ONECUT2 | 10346 | 10072 | -0.000 | -0.3825 | No | ||
| 63 | IL22 | 11948 | 10124 | -0.000 | -0.3853 | No | ||
| 64 | HECTD2 | 23876 3721 | 10346 | -0.001 | -0.3972 | No | ||
| 65 | FABP2 | 15430 | 10421 | -0.001 | -0.4011 | No | ||
| 66 | NCDN | 15756 2487 6471 | 10452 | -0.001 | -0.4027 | No | ||
| 67 | OGN | 21642 3215 | 10508 | -0.001 | -0.4056 | No | ||
| 68 | EHF | 4657 | 10541 | -0.001 | -0.4073 | No | ||
| 69 | USP2 | 19480 3052 3043 | 10584 | -0.002 | -0.4095 | No | ||
| 70 | LGI1 | 23867 | 10920 | -0.002 | -0.4275 | No | ||
| 71 | CASQ2 | 15477 | 11315 | -0.003 | -0.4487 | No | ||
| 72 | RTN1 | 4177 2099 | 11694 | -0.005 | -0.4689 | No | ||
| 73 | MSX2 | 21461 | 11708 | -0.005 | -0.4695 | No | ||
| 74 | SOX5 | 9850 1044 5480 16937 | 11802 | -0.005 | -0.4743 | No | ||
| 75 | UBXD3 | 15701 | 11966 | -0.005 | -0.4829 | No | ||
| 76 | ROS1 | 19771 | 11984 | -0.006 | -0.4836 | No | ||
| 77 | ACCN1 | 20327 1194 | 12053 | -0.006 | -0.4870 | No | ||
| 78 | SIX1 | 9821 | 12055 | -0.006 | -0.4868 | No | ||
| 79 | SATB2 | 5604 | 12242 | -0.007 | -0.4966 | No | ||
| 80 | TSHB | 15218 | 12383 | -0.007 | -0.5039 | No | ||
| 81 | ESRRG | 6447 | 12405 | -0.007 | -0.5047 | No | ||
| 82 | IL7 | 4921 | 12843 | -0.009 | -0.5280 | No | ||
| 83 | HIPK1 | 4851 | 12958 | -0.010 | -0.5337 | No | ||
| 84 | POU4F2 | 9603 5276 | 13020 | -0.010 | -0.5366 | No | ||
| 85 | BACE2 | 1738 22687 1695 | 13152 | -0.011 | -0.5432 | No | ||
| 86 | PTGFR | 15138 | 13198 | -0.011 | -0.5452 | No | ||
| 87 | HOXB8 | 9111 | 13390 | -0.013 | -0.5550 | No | ||
| 88 | CSMD3 | 10735 7938 | 13543 | -0.014 | -0.5627 | No | ||
| 89 | PLAG1 | 7148 15949 12161 | 13585 | -0.014 | -0.5643 | No | ||
| 90 | A2BP1 | 11212 1652 1689 | 13682 | -0.015 | -0.5689 | No | ||
| 91 | SORCS1 | 7184 12203 | 13755 | -0.016 | -0.5721 | No | ||
| 92 | HTR3B | 19129 3145 | 13872 | -0.017 | -0.5776 | No | ||
| 93 | MGLL | 17368 1145 | 14017 | -0.019 | -0.5846 | No | ||
| 94 | SLC26A9 | 11560 980 4067 | 14192 | -0.022 | -0.5931 | No | ||
| 95 | XYLT2 | 20288 | 14200 | -0.022 | -0.5926 | No | ||
| 96 | UBE2R2 | 16240 | 14242 | -0.023 | -0.5939 | No | ||
| 97 | HSPB2 | 19121 | 14454 | -0.028 | -0.6041 | No | ||
| 98 | TMOD4 | 1816 1884 15509 | 14524 | -0.029 | -0.6067 | No | ||
| 99 | DMD | 24295 2647 | 14603 | -0.032 | -0.6096 | No | ||
| 100 | NF1 | 5165 | 15715 | -0.100 | -0.6656 | No | ||
| 101 | SRPK2 | 5513 | 15728 | -0.101 | -0.6621 | No | ||
| 102 | SESN3 | 19561 | 15766 | -0.106 | -0.6598 | No | ||
| 103 | PPARGC1A | 16533 | 15834 | -0.119 | -0.6586 | No | ||
| 104 | FUT11 | 13084 | 15848 | -0.120 | -0.6544 | No | ||
| 105 | CPNE1 | 11186 | 16035 | -0.154 | -0.6581 | No | ||
| 106 | ARHGDIB | 16950 | 16436 | -0.256 | -0.6693 | Yes | ||
| 107 | ZFPM1 | 18439 | 16600 | -0.306 | -0.6656 | Yes | ||
| 108 | WDR23 | 22009 | 16795 | -0.371 | -0.6609 | Yes | ||
| 109 | CRYAB | 19457 | 17296 | -0.572 | -0.6646 | Yes | ||
| 110 | MEF2A | 1201 5089 3492 | 17426 | -0.654 | -0.6448 | Yes | ||
| 111 | SMAD5 | 21621 | 17474 | -0.687 | -0.6193 | Yes | ||
| 112 | ACTR1A | 23653 | 17500 | -0.703 | -0.5919 | Yes | ||
| 113 | TOB1 | 20703 | 17648 | -0.791 | -0.5676 | Yes | ||
| 114 | MBNL1 | 1921 15582 | 17894 | -1.001 | -0.5399 | Yes | ||
| 115 | LUC7L | 47 1520 | 18001 | -1.106 | -0.5005 | Yes | ||
| 116 | RHOG | 17727 | 18012 | -1.123 | -0.4552 | Yes | ||
| 117 | CHD2 | 10847 | 18049 | -1.174 | -0.4092 | Yes | ||
| 118 | MBNL2 | 21932 | 18123 | -1.268 | -0.3613 | Yes | ||
| 119 | ARHGAP8 | 4268 2196 | 18132 | -1.283 | -0.3093 | Yes | ||
| 120 | RAI1 | 5355 20861 | 18227 | -1.454 | -0.2550 | Yes | ||
| 121 | HHEX | 23872 | 18228 | -1.455 | -0.1956 | Yes | ||
| 122 | HIST1H4C | 6643 | 18262 | -1.537 | -0.1346 | Yes | ||
| 123 | LMO2 | 9281 | 18578 | -3.763 | 0.0021 | Yes |

