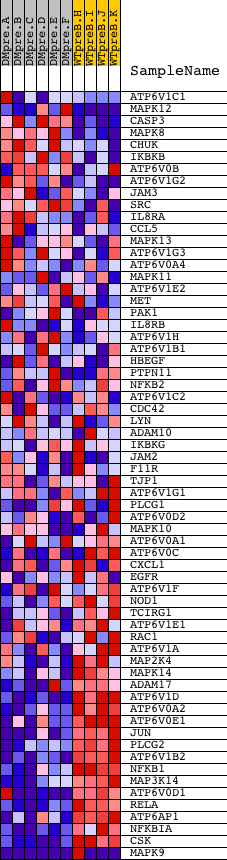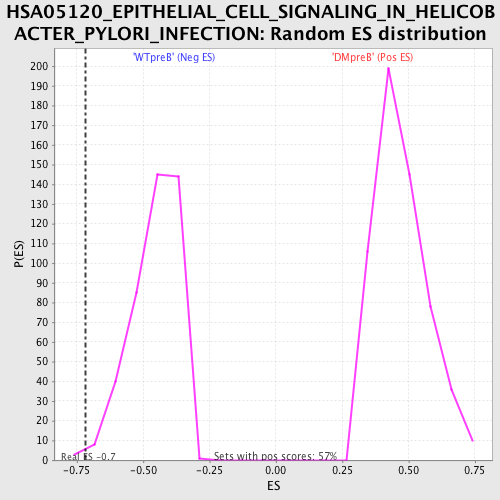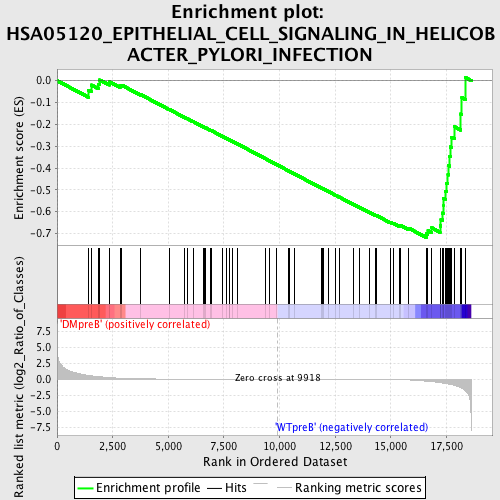
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | HSA05120_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION |
| Enrichment Score (ES) | -0.7180774 |
| Normalized Enrichment Score (NES) | -1.5700614 |
| Nominal p-value | 0.0070422534 |
| FDR q-value | 0.078790806 |
| FWER p-Value | 0.46 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP6V1C1 | 22487 12329 | 1403 | 0.586 | -0.0426 | No | ||
| 2 | MAPK12 | 2182 2215 22164 | 1533 | 0.532 | -0.0196 | No | ||
| 3 | CASP3 | 8693 | 1868 | 0.395 | -0.0154 | No | ||
| 4 | MAPK8 | 6459 | 1918 | 0.379 | 0.0033 | No | ||
| 5 | CHUK | 23665 | 2349 | 0.253 | -0.0056 | No | ||
| 6 | IKBKB | 4907 | 2832 | 0.138 | -0.0238 | No | ||
| 7 | ATP6V0B | 15786 | 2902 | 0.126 | -0.0204 | No | ||
| 8 | ATP6V1G2 | 23257 1534 | 3760 | 0.050 | -0.0638 | No | ||
| 9 | JAM3 | 8197 | 5059 | 0.020 | -0.1326 | No | ||
| 10 | SRC | 5507 | 5732 | 0.014 | -0.1681 | No | ||
| 11 | IL8RA | 13925 | 5870 | 0.013 | -0.1747 | No | ||
| 12 | CCL5 | 20320 | 6124 | 0.012 | -0.1877 | No | ||
| 13 | MAPK13 | 23314 | 6574 | 0.010 | -0.2114 | No | ||
| 14 | ATP6V1G3 | 14107 | 6627 | 0.009 | -0.2136 | No | ||
| 15 | ATP6V0A4 | 8934 | 6690 | 0.009 | -0.2165 | No | ||
| 16 | MAPK11 | 2264 9618 22163 | 6874 | 0.008 | -0.2258 | No | ||
| 17 | ATP6V1E2 | 22880 | 6941 | 0.008 | -0.2289 | No | ||
| 18 | MET | 17520 | 7437 | 0.006 | -0.2553 | No | ||
| 19 | PAK1 | 9527 | 7592 | 0.006 | -0.2632 | No | ||
| 20 | IL8RB | 8752 4533 | 7746 | 0.005 | -0.2712 | No | ||
| 21 | ATP6V1H | 14301 | 7766 | 0.005 | -0.2719 | No | ||
| 22 | ATP6V1B1 | 8499 | 7902 | 0.005 | -0.2789 | No | ||
| 23 | HBEGF | 23457 | 8119 | 0.004 | -0.2903 | No | ||
| 24 | PTPN11 | 5326 16391 9660 | 9360 | 0.001 | -0.3571 | No | ||
| 25 | NFKB2 | 23810 | 9535 | 0.001 | -0.3664 | No | ||
| 26 | ATP6V1C2 | 21098 | 9870 | 0.000 | -0.3844 | No | ||
| 27 | CDC42 | 4503 8722 4504 2465 | 10407 | -0.001 | -0.4132 | No | ||
| 28 | LYN | 16281 | 10463 | -0.001 | -0.4161 | No | ||
| 29 | ADAM10 | 4336 | 10680 | -0.002 | -0.4277 | No | ||
| 30 | IKBKG | 2570 2562 4908 | 11885 | -0.005 | -0.4923 | No | ||
| 31 | JAM2 | 7428 22718 12519 | 11945 | -0.005 | -0.4951 | No | ||
| 32 | F11R | 9199 | 11952 | -0.005 | -0.4952 | No | ||
| 33 | TJP1 | 17803 | 12207 | -0.006 | -0.5085 | No | ||
| 34 | ATP6V1G1 | 12322 | 12525 | -0.008 | -0.5251 | No | ||
| 35 | PLCG1 | 14753 | 12673 | -0.009 | -0.5326 | No | ||
| 36 | ATP6V0D2 | 15934 | 13322 | -0.012 | -0.5668 | No | ||
| 37 | MAPK10 | 11169 | 13577 | -0.014 | -0.5797 | No | ||
| 38 | ATP6V0A1 | 8640 4424 1197 | 14042 | -0.020 | -0.6036 | No | ||
| 39 | ATP6V0C | 8643 | 14304 | -0.025 | -0.6163 | No | ||
| 40 | CXCL1 | 9044 | 14336 | -0.025 | -0.6165 | No | ||
| 41 | EGFR | 1329 20944 | 14989 | -0.045 | -0.6491 | No | ||
| 42 | ATP6V1F | 12291 | 15125 | -0.052 | -0.6535 | No | ||
| 43 | NOD1 | 17141 | 15383 | -0.068 | -0.6635 | No | ||
| 44 | TCIRG1 | 3700 3752 23951 | 15423 | -0.070 | -0.6617 | No | ||
| 45 | ATP6V1E1 | 4423 | 15814 | -0.115 | -0.6762 | No | ||
| 46 | RAC1 | 16302 | 16592 | -0.304 | -0.7010 | Yes | ||
| 47 | ATP6V1A | 8638 | 16670 | -0.331 | -0.6865 | Yes | ||
| 48 | MAP2K4 | 20405 | 16842 | -0.384 | -0.6741 | Yes | ||
| 49 | MAPK14 | 23313 | 17215 | -0.531 | -0.6642 | Yes | ||
| 50 | ADAM17 | 4343 | 17253 | -0.549 | -0.6353 | Yes | ||
| 51 | ATP6V1D | 7872 | 17322 | -0.586 | -0.6059 | Yes | ||
| 52 | ATP6V0A2 | 5760 | 17354 | -0.605 | -0.5735 | Yes | ||
| 53 | ATP6V0E1 | 23326 | 17360 | -0.610 | -0.5395 | Yes | ||
| 54 | JUN | 15832 | 17458 | -0.677 | -0.5066 | Yes | ||
| 55 | PLCG2 | 18453 | 17511 | -0.711 | -0.4693 | Yes | ||
| 56 | ATP6V1B2 | 18599 | 17568 | -0.741 | -0.4307 | Yes | ||
| 57 | NFKB1 | 15160 | 17596 | -0.757 | -0.3895 | Yes | ||
| 58 | MAP3K14 | 11998 | 17649 | -0.792 | -0.3477 | Yes | ||
| 59 | ATP6V0D1 | 3895 8639 3920 | 17671 | -0.807 | -0.3034 | Yes | ||
| 60 | RELA | 23783 | 17734 | -0.859 | -0.2583 | Yes | ||
| 61 | ATP6AP1 | 24296 | 17855 | -0.965 | -0.2104 | Yes | ||
| 62 | NFKBIA | 21065 | 18148 | -1.309 | -0.1524 | Yes | ||
| 63 | CSK | 8805 | 18193 | -1.378 | -0.0772 | Yes | ||
| 64 | MAPK9 | 1233 20903 1383 | 18352 | -1.775 | 0.0142 | Yes |