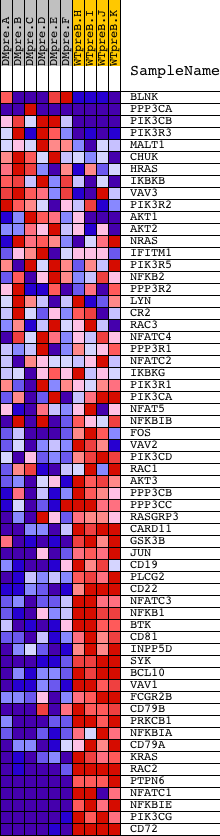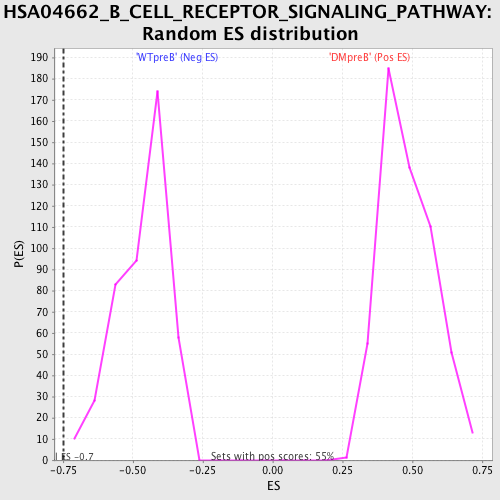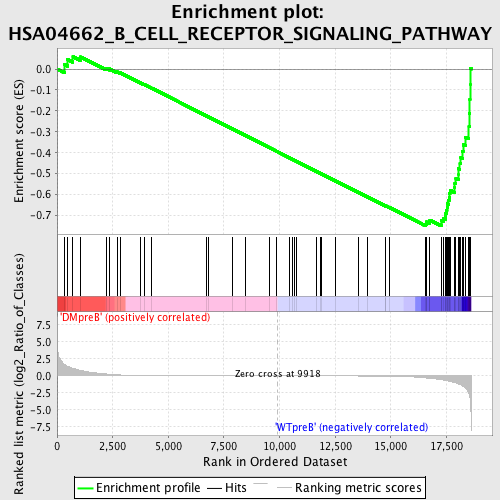
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | HSA04662_B_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.7498295 |
| Normalized Enrichment Score (NES) | -1.6116992 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07877508 |
| FWER p-Value | 0.225 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BLNK | 23681 3691 | 325 | 1.702 | 0.0215 | No | ||
| 2 | PPP3CA | 1863 5284 | 472 | 1.412 | 0.0460 | No | ||
| 3 | PIK3CB | 19030 | 713 | 1.094 | 0.0582 | No | ||
| 4 | PIK3R3 | 5248 | 1062 | 0.797 | 0.0577 | No | ||
| 5 | MALT1 | 6274 | 2207 | 0.289 | 0.0027 | No | ||
| 6 | CHUK | 23665 | 2349 | 0.253 | 0.0009 | No | ||
| 7 | HRAS | 4868 | 2695 | 0.167 | -0.0139 | No | ||
| 8 | IKBKB | 4907 | 2832 | 0.138 | -0.0180 | No | ||
| 9 | VAV3 | 1774 1848 15443 | 3728 | 0.051 | -0.0651 | No | ||
| 10 | PIK3R2 | 18850 | 3916 | 0.043 | -0.0742 | No | ||
| 11 | AKT1 | 8568 | 4238 | 0.034 | -0.0907 | No | ||
| 12 | AKT2 | 4365 4366 | 6731 | 0.009 | -0.2248 | No | ||
| 13 | NRAS | 5191 | 6782 | 0.009 | -0.2273 | No | ||
| 14 | IFITM1 | 18017 7578 12724 | 7895 | 0.005 | -0.2871 | No | ||
| 15 | PIK3R5 | 11507 | 8445 | 0.003 | -0.3166 | No | ||
| 16 | NFKB2 | 23810 | 9535 | 0.001 | -0.3753 | No | ||
| 17 | PPP3R2 | 9612 | 9858 | 0.000 | -0.3927 | No | ||
| 18 | LYN | 16281 | 10463 | -0.001 | -0.4252 | No | ||
| 19 | CR2 | 3942 13707 | 10585 | -0.002 | -0.4317 | No | ||
| 20 | RAC3 | 20561 | 10656 | -0.002 | -0.4354 | No | ||
| 21 | NFATC4 | 22002 | 10757 | -0.002 | -0.4407 | No | ||
| 22 | PPP3R1 | 9611 489 | 11660 | -0.005 | -0.4893 | No | ||
| 23 | NFATC2 | 5168 2866 | 11816 | -0.005 | -0.4975 | No | ||
| 24 | IKBKG | 2570 2562 4908 | 11885 | -0.005 | -0.5010 | No | ||
| 25 | PIK3R1 | 3170 | 12501 | -0.008 | -0.5340 | No | ||
| 26 | PIK3CA | 9562 | 13536 | -0.014 | -0.5894 | No | ||
| 27 | NFAT5 | 3921 7037 12036 | 13952 | -0.019 | -0.6114 | No | ||
| 28 | NFKBIB | 17906 | 14759 | -0.037 | -0.6539 | No | ||
| 29 | FOS | 21202 | 14780 | -0.038 | -0.6542 | No | ||
| 30 | VAV2 | 5848 2670 | 14953 | -0.044 | -0.6624 | No | ||
| 31 | PIK3CD | 9563 | 16569 | -0.297 | -0.7427 | Yes | ||
| 32 | RAC1 | 16302 | 16592 | -0.304 | -0.7369 | Yes | ||
| 33 | AKT3 | 13739 982 | 16609 | -0.309 | -0.7306 | Yes | ||
| 34 | PPP3CB | 5285 | 16735 | -0.352 | -0.7293 | Yes | ||
| 35 | PPP3CC | 21763 | 16753 | -0.357 | -0.7220 | Yes | ||
| 36 | RASGRP3 | 10767 | 17270 | -0.560 | -0.7370 | Yes | ||
| 37 | CARD11 | 8436 | 17271 | -0.561 | -0.7241 | Yes | ||
| 38 | GSK3B | 22761 | 17375 | -0.618 | -0.7155 | Yes | ||
| 39 | JUN | 15832 | 17458 | -0.677 | -0.7044 | Yes | ||
| 40 | CD19 | 17640 | 17470 | -0.683 | -0.6893 | Yes | ||
| 41 | PLCG2 | 18453 | 17511 | -0.711 | -0.6752 | Yes | ||
| 42 | CD22 | 17881 | 17526 | -0.717 | -0.6595 | Yes | ||
| 43 | NFATC3 | 5169 | 17539 | -0.726 | -0.6435 | Yes | ||
| 44 | NFKB1 | 15160 | 17596 | -0.757 | -0.6291 | Yes | ||
| 45 | BTK | 24061 | 17622 | -0.771 | -0.6128 | Yes | ||
| 46 | CD81 | 8719 | 17641 | -0.785 | -0.5958 | Yes | ||
| 47 | INPP5D | 14198 | 17662 | -0.800 | -0.5785 | Yes | ||
| 48 | SYK | 21636 | 17870 | -0.978 | -0.5672 | Yes | ||
| 49 | BCL10 | 15397 | 17878 | -0.984 | -0.5450 | Yes | ||
| 50 | VAV1 | 23173 | 17925 | -1.033 | -0.5238 | Yes | ||
| 51 | FCGR2B | 8959 | 18041 | -1.163 | -0.5033 | Yes | ||
| 52 | CD79B | 20185 1309 | 18046 | -1.168 | -0.4767 | Yes | ||
| 53 | PRKCB1 | 1693 9574 | 18108 | -1.252 | -0.4513 | Yes | ||
| 54 | NFKBIA | 21065 | 18148 | -1.309 | -0.4234 | Yes | ||
| 55 | CD79A | 18342 | 18218 | -1.423 | -0.3945 | Yes | ||
| 56 | KRAS | 9247 | 18253 | -1.521 | -0.3614 | Yes | ||
| 57 | RAC2 | 22217 | 18359 | -1.796 | -0.3259 | Yes | ||
| 58 | PTPN6 | 17002 | 18512 | -2.552 | -0.2755 | Yes | ||
| 59 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 18532 | -2.758 | -0.2133 | Yes | ||
| 60 | NFKBIE | 23225 1556 | 18553 | -3.039 | -0.1446 | Yes | ||
| 61 | PIK3CG | 6635 | 18560 | -3.168 | -0.0723 | Yes | ||
| 62 | CD72 | 8718 | 18566 | -3.281 | 0.0027 | Yes |