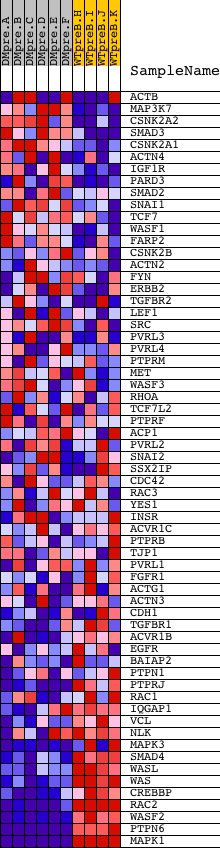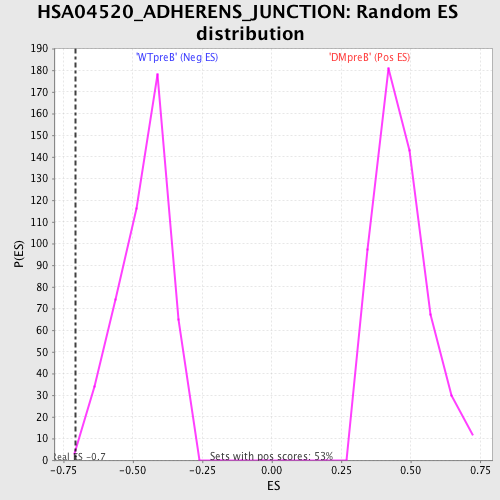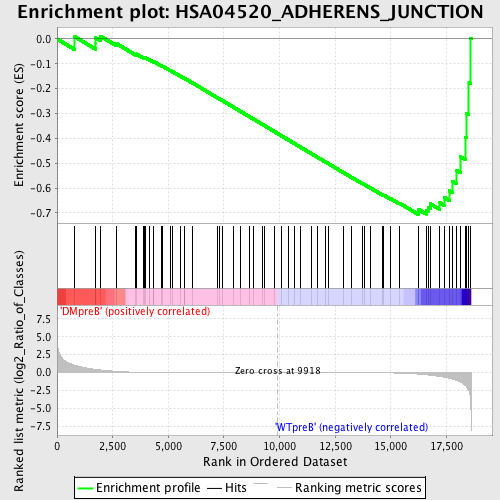
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | HSA04520_ADHERENS_JUNCTION |
| Enrichment Score (ES) | -0.70606554 |
| Normalized Enrichment Score (NES) | -1.5293607 |
| Nominal p-value | 0.0021276595 |
| FDR q-value | 0.10238306 |
| FWER p-Value | 0.748 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ACTB | 8534 337 337 338 | 798 | 1.023 | 0.0091 | No | ||
| 2 | MAP3K7 | 16255 | 1728 | 0.446 | -0.0183 | No | ||
| 3 | CSNK2A2 | 4567 8808 | 1730 | 0.445 | 0.0043 | No | ||
| 4 | SMAD3 | 19084 | 1947 | 0.370 | 0.0115 | No | ||
| 5 | CSNK2A1 | 14797 | 2669 | 0.175 | -0.0184 | No | ||
| 6 | ACTN4 | 17905 1798 1983 | 3538 | 0.060 | -0.0622 | No | ||
| 7 | IGF1R | 9157 | 3561 | 0.059 | -0.0603 | No | ||
| 8 | PARD3 | 8212 13553 | 3870 | 0.045 | -0.0746 | No | ||
| 9 | SMAD2 | 23511 | 3937 | 0.043 | -0.0760 | No | ||
| 10 | SNAI1 | 14726 | 3955 | 0.042 | -0.0748 | No | ||
| 11 | TCF7 | 1467 20466 | 4160 | 0.036 | -0.0839 | No | ||
| 12 | WASF1 | 13514 | 4334 | 0.032 | -0.0917 | No | ||
| 13 | FARP2 | 14175 | 4342 | 0.032 | -0.0904 | No | ||
| 14 | CSNK2B | 23008 | 4678 | 0.025 | -0.1072 | No | ||
| 15 | ACTN2 | 21545 | 4737 | 0.024 | -0.1091 | No | ||
| 16 | FYN | 3375 3395 20052 | 5080 | 0.020 | -0.1265 | No | ||
| 17 | ERBB2 | 8913 | 5205 | 0.019 | -0.1322 | No | ||
| 18 | TGFBR2 | 2994 3001 10167 | 5531 | 0.016 | -0.1489 | No | ||
| 19 | LEF1 | 1860 15420 | 5729 | 0.014 | -0.1588 | No | ||
| 20 | SRC | 5507 | 5732 | 0.014 | -0.1582 | No | ||
| 21 | PVRL3 | 1674 22579 1675 | 6069 | 0.012 | -0.1757 | No | ||
| 22 | PVRL4 | 14055 970 | 7191 | 0.007 | -0.2358 | No | ||
| 23 | PTPRM | 22904 | 7318 | 0.007 | -0.2422 | No | ||
| 24 | MET | 17520 | 7437 | 0.006 | -0.2483 | No | ||
| 25 | WASF3 | 16625 | 7943 | 0.005 | -0.2752 | No | ||
| 26 | RHOA | 8624 4409 4410 | 8262 | 0.004 | -0.2922 | No | ||
| 27 | TCF7L2 | 10048 5646 | 8665 | 0.003 | -0.3137 | No | ||
| 28 | PTPRF | 5331 2545 | 8807 | 0.003 | -0.3211 | No | ||
| 29 | ACP1 | 4317 | 9230 | 0.002 | -0.3438 | No | ||
| 30 | PVRL2 | 9671 5336 | 9310 | 0.001 | -0.3480 | No | ||
| 31 | SNAI2 | 22848 | 9750 | 0.000 | -0.3716 | No | ||
| 32 | SSX2IP | 13617 | 10079 | -0.000 | -0.3893 | No | ||
| 33 | CDC42 | 4503 8722 4504 2465 | 10407 | -0.001 | -0.4069 | No | ||
| 34 | RAC3 | 20561 | 10656 | -0.002 | -0.4201 | No | ||
| 35 | YES1 | 5930 | 10960 | -0.003 | -0.4363 | No | ||
| 36 | INSR | 18950 | 11422 | -0.004 | -0.4610 | No | ||
| 37 | ACVR1C | 14585 | 11711 | -0.005 | -0.4763 | No | ||
| 38 | PTPRB | 19875 | 12069 | -0.006 | -0.4952 | No | ||
| 39 | TJP1 | 17803 | 12207 | -0.006 | -0.5023 | No | ||
| 40 | PVRL1 | 3006 19482 12211 7192 | 12853 | -0.009 | -0.5366 | No | ||
| 41 | FGFR1 | 3789 8968 | 13224 | -0.012 | -0.5559 | No | ||
| 42 | ACTG1 | 8535 | 13713 | -0.016 | -0.5814 | No | ||
| 43 | ACTN3 | 23967 8540 | 13818 | -0.017 | -0.5862 | No | ||
| 44 | CDH1 | 18479 | 14069 | -0.020 | -0.5986 | No | ||
| 45 | TGFBR1 | 16213 10165 5745 | 14629 | -0.032 | -0.6271 | No | ||
| 46 | ACVR1B | 4335 22353 | 14681 | -0.034 | -0.6281 | No | ||
| 47 | EGFR | 1329 20944 | 14989 | -0.045 | -0.6424 | No | ||
| 48 | BAIAP2 | 449 4236 | 15384 | -0.068 | -0.6601 | No | ||
| 49 | PTPN1 | 5325 | 16237 | -0.200 | -0.6959 | Yes | ||
| 50 | PTPRJ | 9664 | 16260 | -0.207 | -0.6865 | Yes | ||
| 51 | RAC1 | 16302 | 16592 | -0.304 | -0.6889 | Yes | ||
| 52 | IQGAP1 | 6619 | 16674 | -0.333 | -0.6763 | Yes | ||
| 53 | VCL | 22083 | 16779 | -0.367 | -0.6633 | Yes | ||
| 54 | NLK | 5179 5178 | 17203 | -0.527 | -0.6592 | Yes | ||
| 55 | MAPK3 | 6458 11170 | 17396 | -0.634 | -0.6373 | Yes | ||
| 56 | SMAD4 | 5058 | 17630 | -0.777 | -0.6103 | Yes | ||
| 57 | WASL | 17203 | 17758 | -0.876 | -0.5726 | Yes | ||
| 58 | WAS | 24193 | 17956 | -1.065 | -0.5290 | Yes | ||
| 59 | CREBBP | 22682 8783 | 18124 | -1.271 | -0.4733 | Yes | ||
| 60 | RAC2 | 22217 | 18359 | -1.796 | -0.3944 | Yes | ||
| 61 | WASF2 | 6326 | 18391 | -1.916 | -0.2985 | Yes | ||
| 62 | PTPN6 | 17002 | 18512 | -2.552 | -0.1751 | Yes | ||
| 63 | MAPK1 | 1642 11167 | 18573 | -3.548 | 0.0023 | Yes |