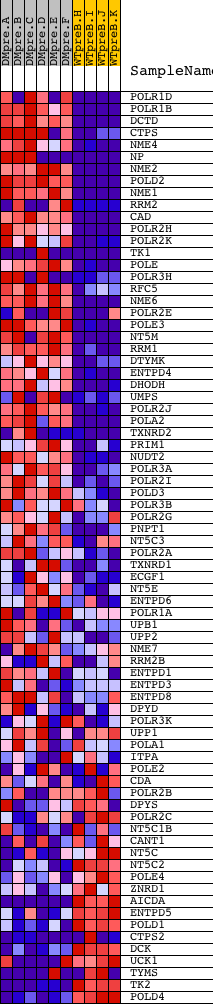
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | HSA00240_PYRIMIDINE_METABOLISM |
| Enrichment Score (ES) | 0.7374033 |
| Normalized Enrichment Score (NES) | 1.5989481 |
| Nominal p-value | 0.0017636684 |
| FDR q-value | 0.05829073 |
| FWER p-Value | 0.202 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POLR1D | 3593 3658 16623 | 4 | 5.920 | 0.0884 | Yes | ||
| 2 | POLR1B | 14857 | 36 | 3.607 | 0.1407 | Yes | ||
| 3 | DCTD | 18620 | 38 | 3.458 | 0.1924 | Yes | ||
| 4 | CTPS | 2514 15772 | 86 | 2.716 | 0.2306 | Yes | ||
| 5 | NME4 | 23065 1577 | 120 | 2.545 | 0.2669 | Yes | ||
| 6 | NP | 22027 9597 5273 5274 | 123 | 2.541 | 0.3048 | Yes | ||
| 7 | NME2 | 9468 | 150 | 2.439 | 0.3399 | Yes | ||
| 8 | POLD2 | 20537 | 230 | 2.045 | 0.3663 | Yes | ||
| 9 | NME1 | 9467 | 273 | 1.870 | 0.3920 | Yes | ||
| 10 | RRM2 | 5401 5400 | 303 | 1.762 | 0.4168 | Yes | ||
| 11 | CAD | 16886 | 321 | 1.716 | 0.4416 | Yes | ||
| 12 | POLR2H | 10888 | 365 | 1.631 | 0.4637 | Yes | ||
| 13 | POLR2K | 9413 | 407 | 1.525 | 0.4843 | Yes | ||
| 14 | TK1 | 1457 10182 5762 | 442 | 1.459 | 0.5043 | Yes | ||
| 15 | POLE | 16755 | 455 | 1.444 | 0.5252 | Yes | ||
| 16 | POLR3H | 13460 8135 | 471 | 1.412 | 0.5456 | Yes | ||
| 17 | RFC5 | 13005 7791 | 491 | 1.389 | 0.5653 | Yes | ||
| 18 | NME6 | 3139 19297 | 523 | 1.347 | 0.5838 | Yes | ||
| 19 | POLR2E | 3325 19699 | 551 | 1.316 | 0.6021 | Yes | ||
| 20 | POLE3 | 7200 | 650 | 1.174 | 0.6143 | Yes | ||
| 21 | NT5M | 8345 4175 | 671 | 1.154 | 0.6305 | Yes | ||
| 22 | RRM1 | 18163 | 718 | 1.089 | 0.6444 | Yes | ||
| 23 | DTYMK | 5776 | 720 | 1.087 | 0.6606 | Yes | ||
| 24 | ENTPD4 | 7449 | 742 | 1.070 | 0.6755 | Yes | ||
| 25 | DHODH | 7152 | 801 | 1.020 | 0.6876 | Yes | ||
| 26 | UMPS | 22606 1760 | 1006 | 0.838 | 0.6891 | Yes | ||
| 27 | POLR2J | 16672 | 1011 | 0.836 | 0.7014 | Yes | ||
| 28 | POLA2 | 23988 | 1088 | 0.782 | 0.7090 | Yes | ||
| 29 | TXNRD2 | 22829 1649 | 1135 | 0.753 | 0.7178 | Yes | ||
| 30 | PRIM1 | 19847 | 1166 | 0.734 | 0.7272 | Yes | ||
| 31 | NUDT2 | 16239 | 1179 | 0.724 | 0.7374 | Yes | ||
| 32 | POLR3A | 21900 | 1748 | 0.438 | 0.7133 | No | ||
| 33 | POLR2I | 12839 | 1841 | 0.404 | 0.7144 | No | ||
| 34 | POLD3 | 17742 | 2092 | 0.323 | 0.7058 | No | ||
| 35 | POLR3B | 12875 | 2603 | 0.192 | 0.6811 | No | ||
| 36 | POLR2G | 23753 | 2623 | 0.184 | 0.6828 | No | ||
| 37 | PNPT1 | 20936 | 2683 | 0.171 | 0.6822 | No | ||
| 38 | NT5C3 | 17136 | 2720 | 0.162 | 0.6827 | No | ||
| 39 | POLR2A | 5394 | 2733 | 0.159 | 0.6844 | No | ||
| 40 | TXNRD1 | 19923 | 2839 | 0.137 | 0.6808 | No | ||
| 41 | ECGF1 | 22160 | 2875 | 0.132 | 0.6809 | No | ||
| 42 | NT5E | 19360 18702 | 3615 | 0.057 | 0.6419 | No | ||
| 43 | ENTPD6 | 4496 2678 | 4455 | 0.029 | 0.5971 | No | ||
| 44 | POLR1A | 9749 5393 | 4624 | 0.026 | 0.5884 | No | ||
| 45 | UPB1 | 19988 3389 | 4796 | 0.023 | 0.5795 | No | ||
| 46 | UPP2 | 15009 13346 8032 | 5341 | 0.018 | 0.5505 | No | ||
| 47 | NME7 | 926 4101 14070 3986 3946 931 | 5926 | 0.013 | 0.5192 | No | ||
| 48 | RRM2B | 6902 6903 11801 | 6266 | 0.011 | 0.5010 | No | ||
| 49 | ENTPD1 | 4495 | 6431 | 0.010 | 0.4923 | No | ||
| 50 | ENTPD3 | 19266 | 6632 | 0.009 | 0.4817 | No | ||
| 51 | ENTPD8 | 12996 | 6756 | 0.009 | 0.4752 | No | ||
| 52 | DPYD | 15437 | 6866 | 0.008 | 0.4694 | No | ||
| 53 | POLR3K | 12447 7372 | 8352 | 0.004 | 0.3894 | No | ||
| 54 | UPP1 | 20947 1385 | 9018 | 0.002 | 0.3536 | No | ||
| 55 | POLA1 | 24112 | 9841 | 0.000 | 0.3092 | No | ||
| 56 | ITPA | 9193 | 10238 | -0.001 | 0.2879 | No | ||
| 57 | POLE2 | 21053 | 10899 | -0.002 | 0.2523 | No | ||
| 58 | CDA | 15702 | 11081 | -0.003 | 0.2426 | No | ||
| 59 | POLR2B | 16817 | 11867 | -0.005 | 0.2003 | No | ||
| 60 | DPYS | 22305 | 12213 | -0.006 | 0.1818 | No | ||
| 61 | POLR2C | 9750 | 12996 | -0.010 | 0.1398 | No | ||
| 62 | NT5C1B | 21315 | 14335 | -0.025 | 0.0680 | No | ||
| 63 | CANT1 | 13304 | 14557 | -0.030 | 0.0566 | No | ||
| 64 | NT5C | 20151 | 16651 | -0.327 | -0.0514 | No | ||
| 65 | NT5C2 | 3768 8052 | 16811 | -0.374 | -0.0544 | No | ||
| 66 | POLE4 | 12441 | 17037 | -0.460 | -0.0597 | No | ||
| 67 | ZNRD1 | 1491 22987 | 17058 | -0.468 | -0.0537 | No | ||
| 68 | AICDA | 17295 1087 | 17149 | -0.509 | -0.0510 | No | ||
| 69 | ENTPD5 | 4497 | 17182 | -0.520 | -0.0449 | No | ||
| 70 | POLD1 | 17847 | 17231 | -0.537 | -0.0395 | No | ||
| 71 | CTPS2 | 12060 2571 | 17449 | -0.671 | -0.0411 | No | ||
| 72 | DCK | 16808 | 17693 | -0.826 | -0.0419 | No | ||
| 73 | UCK1 | 10254 5831 10253 | 17808 | -0.918 | -0.0343 | No | ||
| 74 | TYMS | 5810 5809 3606 3598 | 17913 | -1.023 | -0.0246 | No | ||
| 75 | TK2 | 18779 | 18433 | -2.082 | -0.0214 | No | ||
| 76 | POLD4 | 12822 | 18434 | -2.084 | 0.0098 | No |