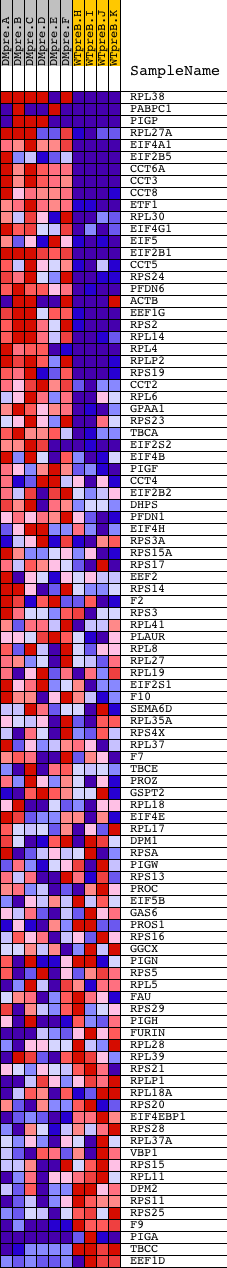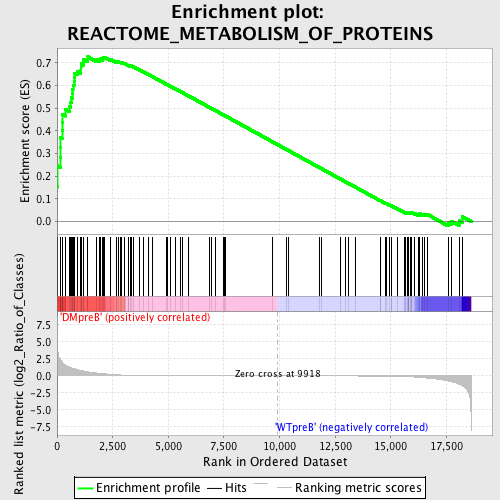
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_DMpreB_versus_WTpreB.phenotype_DMpreB_versus_WTpreB.cls #DMpreB_versus_WTpreB |
| Phenotype | phenotype_DMpreB_versus_WTpreB.cls#DMpreB_versus_WTpreB |
| Upregulated in class | DMpreB |
| GeneSet | REACTOME_METABOLISM_OF_PROTEINS |
| Enrichment Score (ES) | 0.7268911 |
| Normalized Enrichment Score (NES) | 1.6280621 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.02971635 |
| FWER p-Value | 0.151 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL38 | 12562 20606 7475 | 0 | 8.590 | 0.1530 | Yes | ||
| 2 | PABPC1 | 5219 9522 9523 23572 | 7 | 5.273 | 0.2465 | Yes | ||
| 3 | PIGP | 12082 1665 | 137 | 2.497 | 0.2840 | Yes | ||
| 4 | RPL27A | 11181 6467 18130 | 143 | 2.448 | 0.3274 | Yes | ||
| 5 | EIF4A1 | 8889 23719 | 146 | 2.444 | 0.3708 | Yes | ||
| 6 | EIF2B5 | 1719 22822 | 223 | 2.056 | 0.4033 | Yes | ||
| 7 | CCT6A | 16689 | 242 | 1.973 | 0.4375 | Yes | ||
| 8 | CCT3 | 8709 | 248 | 1.945 | 0.4718 | Yes | ||
| 9 | CCT8 | 22551 | 359 | 1.643 | 0.4952 | Yes | ||
| 10 | ETF1 | 23467 | 565 | 1.296 | 0.5072 | Yes | ||
| 11 | RPL30 | 9742 | 610 | 1.225 | 0.5266 | Yes | ||
| 12 | EIF4G1 | 22818 | 635 | 1.192 | 0.5465 | Yes | ||
| 13 | EIF5 | 5736 | 685 | 1.132 | 0.5640 | Yes | ||
| 14 | EIF2B1 | 16368 3458 | 698 | 1.115 | 0.5832 | Yes | ||
| 15 | CCT5 | 22316 | 716 | 1.092 | 0.6018 | Yes | ||
| 16 | RPS24 | 5399 | 764 | 1.046 | 0.6179 | Yes | ||
| 17 | PFDN6 | 23025 | 786 | 1.028 | 0.6350 | Yes | ||
| 18 | ACTB | 8534 337 337 338 | 798 | 1.023 | 0.6527 | Yes | ||
| 19 | EEF1G | 12480 | 904 | 0.920 | 0.6634 | Yes | ||
| 20 | RPS2 | 9279 | 1069 | 0.794 | 0.6686 | Yes | ||
| 21 | RPL14 | 19267 | 1073 | 0.792 | 0.6826 | Yes | ||
| 22 | RPL4 | 7499 19411 | 1082 | 0.785 | 0.6961 | Yes | ||
| 23 | RPLP2 | 7401 | 1183 | 0.722 | 0.7036 | Yes | ||
| 24 | RPS19 | 5398 | 1201 | 0.713 | 0.7154 | Yes | ||
| 25 | CCT2 | 19622 | 1384 | 0.600 | 0.7162 | Yes | ||
| 26 | RPL6 | 9747 | 1385 | 0.599 | 0.7269 | Yes | ||
| 27 | GPAA1 | 22446 | 1773 | 0.429 | 0.7136 | No | ||
| 28 | RPS23 | 12352 | 1899 | 0.385 | 0.7137 | No | ||
| 29 | TBCA | 10037 | 1950 | 0.369 | 0.7176 | No | ||
| 30 | EIF2S2 | 7406 14383 | 2054 | 0.337 | 0.7181 | No | ||
| 31 | EIF4B | 13279 7979 | 2068 | 0.332 | 0.7233 | No | ||
| 32 | PIGF | 22879 | 2144 | 0.306 | 0.7247 | No | ||
| 33 | CCT4 | 8710 | 2386 | 0.242 | 0.7160 | No | ||
| 34 | EIF2B2 | 21204 | 2656 | 0.178 | 0.7046 | No | ||
| 35 | DHPS | 3815 | 2682 | 0.171 | 0.7063 | No | ||
| 36 | PFDN1 | 23458 | 2760 | 0.153 | 0.7049 | No | ||
| 37 | EIF4H | 16352 | 2833 | 0.138 | 0.7034 | No | ||
| 38 | RPS3A | 9755 | 2908 | 0.125 | 0.7017 | No | ||
| 39 | RPS15A | 6476 | 3011 | 0.108 | 0.6981 | No | ||
| 40 | RPS17 | 9753 | 3227 | 0.083 | 0.6880 | No | ||
| 41 | EEF2 | 8881 4654 8882 | 3229 | 0.083 | 0.6894 | No | ||
| 42 | RPS14 | 9751 | 3282 | 0.079 | 0.6880 | No | ||
| 43 | F2 | 14524 | 3326 | 0.074 | 0.6870 | No | ||
| 44 | RPS3 | 6549 11295 | 3415 | 0.067 | 0.6834 | No | ||
| 45 | RPL41 | 12611 | 3724 | 0.051 | 0.6677 | No | ||
| 46 | PLAUR | 18351 | 3886 | 0.044 | 0.6598 | No | ||
| 47 | RPL8 | 22437 | 4085 | 0.038 | 0.6498 | No | ||
| 48 | RPL27 | 9740 | 4272 | 0.033 | 0.6403 | No | ||
| 49 | RPL19 | 9736 | 4906 | 0.022 | 0.6065 | No | ||
| 50 | EIF2S1 | 4658 | 4978 | 0.021 | 0.6031 | No | ||
| 51 | F10 | 18682 | 5081 | 0.020 | 0.5979 | No | ||
| 52 | SEMA6D | 2800 14876 90 | 5340 | 0.018 | 0.5843 | No | ||
| 53 | RPL35A | 12194 | 5559 | 0.016 | 0.5728 | No | ||
| 54 | RPS4X | 9756 | 5617 | 0.015 | 0.5700 | No | ||
| 55 | RPL37 | 12502 7421 22521 | 5908 | 0.013 | 0.5546 | No | ||
| 56 | F7 | 18681 | 6844 | 0.008 | 0.5043 | No | ||
| 57 | TBCE | 21543 | 6922 | 0.008 | 0.5002 | No | ||
| 58 | PROZ | 7360 | 7115 | 0.007 | 0.4900 | No | ||
| 59 | GSPT2 | 4806 | 7481 | 0.006 | 0.4704 | No | ||
| 60 | RPL18 | 450 5390 | 7542 | 0.006 | 0.4673 | No | ||
| 61 | EIF4E | 15403 1827 8890 | 7585 | 0.006 | 0.4651 | No | ||
| 62 | RPL17 | 11429 6653 | 9669 | 0.001 | 0.3526 | No | ||
| 63 | DPM1 | 4636 2882 8860 | 10309 | -0.001 | 0.3181 | No | ||
| 64 | RPSA | 19270 4984 | 10413 | -0.001 | 0.3126 | No | ||
| 65 | PIGW | 20312 | 11815 | -0.005 | 0.2370 | No | ||
| 66 | RPS13 | 12633 | 11876 | -0.005 | 0.2339 | No | ||
| 67 | PROC | 23475 | 12757 | -0.009 | 0.1865 | No | ||
| 68 | EIF5B | 10391 5963 | 12944 | -0.010 | 0.1767 | No | ||
| 69 | GAS6 | 18927 | 13090 | -0.011 | 0.1690 | No | ||
| 70 | PROS1 | 22725 | 13391 | -0.013 | 0.1531 | No | ||
| 71 | RPS16 | 9752 | 14514 | -0.029 | 0.0930 | No | ||
| 72 | GGCX | 17412 992 | 14522 | -0.029 | 0.0931 | No | ||
| 73 | PIGN | 13865 | 14740 | -0.036 | 0.0821 | No | ||
| 74 | RPS5 | 18391 | 14794 | -0.038 | 0.0799 | No | ||
| 75 | RPL5 | 9746 | 14925 | -0.042 | 0.0736 | No | ||
| 76 | FAU | 8954 | 15013 | -0.046 | 0.0697 | No | ||
| 77 | RPS29 | 9754 | 15018 | -0.046 | 0.0703 | No | ||
| 78 | PIGH | 8483 | 15277 | -0.061 | 0.0575 | No | ||
| 79 | FURIN | 9538 | 15620 | -0.089 | 0.0406 | No | ||
| 80 | RPL28 | 5392 | 15681 | -0.095 | 0.0390 | No | ||
| 81 | RPL39 | 12496 | 15742 | -0.102 | 0.0376 | No | ||
| 82 | RPS21 | 12356 | 15776 | -0.108 | 0.0378 | No | ||
| 83 | RPLP1 | 3010 | 15793 | -0.111 | 0.0389 | No | ||
| 84 | RPL18A | 13358 | 15800 | -0.112 | 0.0406 | No | ||
| 85 | RPS20 | 7438 | 15905 | -0.129 | 0.0372 | No | ||
| 86 | EIF4EBP1 | 8891 4661 | 15908 | -0.129 | 0.0394 | No | ||
| 87 | RPS28 | 12009 | 15921 | -0.130 | 0.0411 | No | ||
| 88 | RPL37A | 9744 | 16068 | -0.161 | 0.0361 | No | ||
| 89 | VBP1 | 10281 | 16223 | -0.196 | 0.0313 | No | ||
| 90 | RPS15 | 5396 | 16272 | -0.209 | 0.0324 | No | ||
| 91 | RPL11 | 12450 | 16276 | -0.209 | 0.0360 | No | ||
| 92 | DPM2 | 8861 | 16440 | -0.257 | 0.0318 | No | ||
| 93 | RPS11 | 11317 | 16502 | -0.276 | 0.0334 | No | ||
| 94 | RPS25 | 13270 | 16642 | -0.323 | 0.0316 | No | ||
| 95 | F9 | 24328 | 17584 | -0.750 | -0.0058 | No | ||
| 96 | PIGA | 2592 24210 | 17713 | -0.847 | 0.0023 | No | ||
| 97 | TBCC | 23215 | 18068 | -1.197 | 0.0045 | No | ||
| 98 | EEF1D | 12393 2309 | 18206 | -1.404 | 0.0221 | No |