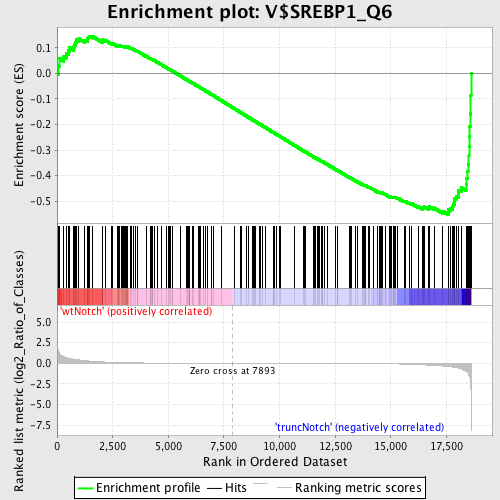
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$SREBP1_Q6 |
| Enrichment Score (ES) | -0.5516919 |
| Normalized Enrichment Score (NES) | -1.427122 |
| Nominal p-value | 0.0080 |
| FDR q-value | 0.63748753 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPP1R10 | 110136 3170725 3610484 | 77 | 1.334 | 0.0294 | No | ||
| 2 | EIF4G2 | 3800575 6860184 | 94 | 1.204 | 0.0588 | No | ||
| 3 | PIAS1 | 5340301 | 305 | 0.780 | 0.0671 | No | ||
| 4 | EMP1 | 4120438 | 432 | 0.662 | 0.0769 | No | ||
| 5 | FCHSD2 | 5720092 | 510 | 0.615 | 0.0882 | No | ||
| 6 | KPNB1 | 1690138 | 543 | 0.602 | 0.1016 | No | ||
| 7 | ATP6V1A | 6590242 | 739 | 0.493 | 0.1035 | No | ||
| 8 | MMP14 | 670079 | 776 | 0.481 | 0.1136 | No | ||
| 9 | NSF | 4070075 | 834 | 0.456 | 0.1220 | No | ||
| 10 | NMT1 | 5810152 | 868 | 0.444 | 0.1314 | No | ||
| 11 | PDCD10 | 4760500 | 982 | 0.404 | 0.1354 | No | ||
| 12 | DDX6 | 2630025 6770408 | 1247 | 0.323 | 0.1293 | No | ||
| 13 | NFIX | 2450152 | 1365 | 0.289 | 0.1302 | No | ||
| 14 | ZFYVE1 | 60440 | 1366 | 0.289 | 0.1375 | No | ||
| 15 | SLIT3 | 7100132 | 1419 | 0.273 | 0.1415 | No | ||
| 16 | ANP32A | 510044 1580095 2680037 | 1462 | 0.264 | 0.1459 | No | ||
| 17 | JUP | 2510671 | 1600 | 0.233 | 0.1444 | No | ||
| 18 | SIPA1L2 | 4850731 | 2031 | 0.163 | 0.1251 | No | ||
| 19 | RBX1 | 2120010 2340047 | 2049 | 0.160 | 0.1282 | No | ||
| 20 | LASP1 | 1690528 2190341 6420341 | 2056 | 0.159 | 0.1319 | No | ||
| 21 | NFAT5 | 2510411 5890195 6550152 | 2188 | 0.140 | 0.1283 | No | ||
| 22 | YWHAQ | 6760524 | 2442 | 0.112 | 0.1174 | No | ||
| 23 | MYH10 | 2640673 | 2489 | 0.107 | 0.1176 | No | ||
| 24 | PHF15 | 6550066 6860673 | 2692 | 0.090 | 0.1089 | No | ||
| 25 | STX1A | 3060273 | 2768 | 0.084 | 0.1070 | No | ||
| 26 | TRERF1 | 6370017 6940138 | 2775 | 0.084 | 0.1088 | No | ||
| 27 | NTRK3 | 4050687 4070167 4610026 6100484 | 2796 | 0.083 | 0.1098 | No | ||
| 28 | FXYD1 | 1400239 6560129 | 2881 | 0.076 | 0.1071 | No | ||
| 29 | DYRK1A | 3190181 | 2922 | 0.073 | 0.1068 | No | ||
| 30 | EN1 | 5360168 7050025 | 2979 | 0.069 | 0.1055 | No | ||
| 31 | MAPKAPK3 | 630520 | 3034 | 0.066 | 0.1042 | No | ||
| 32 | PACSIN3 | 4540369 4540400 5360037 6940129 | 3071 | 0.063 | 0.1039 | No | ||
| 33 | PCF11 | 1240180 | 3114 | 0.061 | 0.1031 | No | ||
| 34 | FBS1 | 2570520 | 3129 | 0.060 | 0.1039 | No | ||
| 35 | B4GALT2 | 780037 | 3164 | 0.057 | 0.1035 | No | ||
| 36 | GPR85 | 5130750 5910020 | 3178 | 0.056 | 0.1042 | No | ||
| 37 | SLC39A14 | 670300 1400093 1990711 | 3304 | 0.050 | 0.0986 | No | ||
| 38 | REL | 360707 | 3352 | 0.048 | 0.0973 | No | ||
| 39 | ZIC4 | 1500082 | 3443 | 0.044 | 0.0935 | No | ||
| 40 | BAZ2A | 730184 | 3525 | 0.041 | 0.0902 | No | ||
| 41 | CLDN5 | 6510717 | 3609 | 0.038 | 0.0866 | No | ||
| 42 | CLASP1 | 6860279 | 3615 | 0.037 | 0.0873 | No | ||
| 43 | SOX12 | 1780037 | 4005 | 0.026 | 0.0668 | No | ||
| 44 | ENO2 | 2320068 | 4193 | 0.023 | 0.0573 | No | ||
| 45 | TRAF4 | 3060041 4920528 6980286 | 4197 | 0.023 | 0.0577 | No | ||
| 46 | DSG4 | 3990605 6400736 | 4254 | 0.022 | 0.0552 | No | ||
| 47 | POU4F2 | 2120195 2570022 | 4288 | 0.021 | 0.0539 | No | ||
| 48 | LRP8 | 3610746 5360035 | 4294 | 0.021 | 0.0542 | No | ||
| 49 | RARG | 6760136 | 4390 | 0.020 | 0.0495 | No | ||
| 50 | SREBF2 | 3390692 | 4495 | 0.018 | 0.0444 | No | ||
| 51 | KIF1B | 1240494 2370139 4570270 6510102 | 4683 | 0.016 | 0.0346 | No | ||
| 52 | TEAD2 | 6450079 | 4929 | 0.013 | 0.0217 | No | ||
| 53 | DHH | 2120433 | 5017 | 0.012 | 0.0172 | No | ||
| 54 | MEOX2 | 6400288 | 5061 | 0.012 | 0.0152 | No | ||
| 55 | KCNN3 | 6520600 6420138 | 5093 | 0.012 | 0.0138 | No | ||
| 56 | SLC4A1 | 6290577 | 5174 | 0.011 | 0.0098 | No | ||
| 57 | CACNA1D | 1740315 4730731 | 5188 | 0.011 | 0.0094 | No | ||
| 58 | SLC4A10 | 2060670 | 5558 | 0.009 | -0.0104 | No | ||
| 59 | LIN28 | 6590672 | 5827 | 0.007 | -0.0248 | No | ||
| 60 | SLC24A1 | 1240463 | 5846 | 0.007 | -0.0256 | No | ||
| 61 | GUCA2B | 4200014 | 5890 | 0.007 | -0.0277 | No | ||
| 62 | TCF2 | 870338 5050632 | 5950 | 0.007 | -0.0308 | No | ||
| 63 | GAP43 | 3130504 | 5970 | 0.007 | -0.0316 | No | ||
| 64 | GRIA1 | 1340152 3780750 4920440 | 5971 | 0.007 | -0.0315 | No | ||
| 65 | GFAP | 2060092 | 6098 | 0.006 | -0.0382 | No | ||
| 66 | GRID2 | 2680242 | 6135 | 0.006 | -0.0400 | No | ||
| 67 | DUSP8 | 6650039 | 6350 | 0.005 | -0.0514 | No | ||
| 68 | EMX2 | 1660092 | 6392 | 0.005 | -0.0535 | No | ||
| 69 | SERPINI1 | 2940468 | 6406 | 0.005 | -0.0541 | No | ||
| 70 | SIX4 | 6760022 | 6443 | 0.005 | -0.0560 | No | ||
| 71 | HYAL1 | 3850341 | 6575 | 0.004 | -0.0630 | No | ||
| 72 | PLA2G3 | 5130739 | 6690 | 0.004 | -0.0691 | No | ||
| 73 | GNAO1 | 2510292 6590487 | 6776 | 0.003 | -0.0736 | No | ||
| 74 | CACNB1 | 2940427 3710487 | 6940 | 0.003 | -0.0824 | No | ||
| 75 | SALL1 | 5420020 7050195 | 6957 | 0.003 | -0.0832 | No | ||
| 76 | SLC2A4 | 540441 | 7048 | 0.002 | -0.0880 | No | ||
| 77 | PAX3 | 50551 | 7370 | 0.001 | -0.1053 | No | ||
| 78 | ZFP36L1 | 2510138 4120048 | 7959 | -0.000 | -0.1372 | No | ||
| 79 | HOXD10 | 6100039 | 8239 | -0.001 | -0.1523 | No | ||
| 80 | KCNMB1 | 4760139 | 8300 | -0.001 | -0.1556 | No | ||
| 81 | ESR1 | 4060372 5860193 | 8509 | -0.002 | -0.1668 | No | ||
| 82 | ANXA9 | 1190195 | 8616 | -0.002 | -0.1725 | No | ||
| 83 | PDE5A | 130072 | 8798 | -0.003 | -0.1822 | No | ||
| 84 | LRRN1 | 3290154 | 8811 | -0.003 | -0.1828 | No | ||
| 85 | TIMP4 | 6110433 | 8886 | -0.003 | -0.1868 | No | ||
| 86 | NF1 | 6980433 | 8907 | -0.003 | -0.1878 | No | ||
| 87 | EVX1 | 1980471 | 9097 | -0.003 | -0.1979 | No | ||
| 88 | NFATC4 | 2470735 | 9153 | -0.004 | -0.2008 | No | ||
| 89 | FABP6 | 7000278 | 9226 | -0.004 | -0.2046 | No | ||
| 90 | LMX1A | 2810239 5690324 | 9369 | -0.004 | -0.2122 | No | ||
| 91 | GREM1 | 3940180 | 9711 | -0.005 | -0.2306 | No | ||
| 92 | HOXA11 | 6980133 | 9741 | -0.005 | -0.2320 | No | ||
| 93 | HIF1A | 5670605 | 9774 | -0.005 | -0.2336 | No | ||
| 94 | FBXO24 | 1230132 4610121 | 9851 | -0.006 | -0.2376 | No | ||
| 95 | ERF | 110551 1770278 | 9994 | -0.006 | -0.2452 | No | ||
| 96 | HOXC4 | 1940193 | 10060 | -0.006 | -0.2485 | No | ||
| 97 | RKHD3 | 6180471 | 10664 | -0.009 | -0.2810 | No | ||
| 98 | FADS3 | 3120079 | 11063 | -0.010 | -0.3023 | No | ||
| 99 | CRLF1 | 3520092 | 11133 | -0.011 | -0.3058 | No | ||
| 100 | PTRF | 2260204 | 11165 | -0.011 | -0.3072 | No | ||
| 101 | ATP6V0C | 1780609 | 11525 | -0.013 | -0.3264 | No | ||
| 102 | AGER | 5860538 5910180 | 11553 | -0.013 | -0.3275 | No | ||
| 103 | EPO | 940180 | 11631 | -0.013 | -0.3313 | No | ||
| 104 | KCNK7 | 4230576 | 11684 | -0.014 | -0.3338 | No | ||
| 105 | MT4 | 4780338 | 11751 | -0.014 | -0.3370 | No | ||
| 106 | DLGAP4 | 4570672 5690161 | 11785 | -0.014 | -0.3385 | No | ||
| 107 | MACF1 | 1340132 2370176 2900750 6590364 | 11879 | -0.015 | -0.3431 | No | ||
| 108 | NEUROD2 | 5290097 | 11907 | -0.015 | -0.3442 | No | ||
| 109 | FLNC | 2360048 5050270 | 11930 | -0.015 | -0.3450 | No | ||
| 110 | SMTN | 670563 1190438 | 12004 | -0.016 | -0.3486 | No | ||
| 111 | LGP2 | 4480338 | 12136 | -0.017 | -0.3553 | No | ||
| 112 | KLF12 | 1660095 4810288 5340546 6520286 | 12491 | -0.020 | -0.3740 | No | ||
| 113 | HOXA3 | 3610632 6020358 | 12594 | -0.021 | -0.3790 | No | ||
| 114 | BCL6B | 60047 | 12596 | -0.021 | -0.3785 | No | ||
| 115 | H2AFY | 4730750 | 13127 | -0.027 | -0.4066 | No | ||
| 116 | INPP4A | 510215 1090541 4850093 6100519 | 13170 | -0.027 | -0.4082 | No | ||
| 117 | TGFB1I1 | 2060288 6550450 | 13215 | -0.028 | -0.4099 | No | ||
| 118 | LRCH4 | 5220279 6510026 | 13424 | -0.031 | -0.4204 | No | ||
| 119 | ABCG4 | 3440458 | 13519 | -0.032 | -0.4247 | No | ||
| 120 | HOXB2 | 6450592 | 13720 | -0.036 | -0.4346 | No | ||
| 121 | PKP3 | 3710048 | 13780 | -0.037 | -0.4369 | No | ||
| 122 | LDLR | 5670386 | 13802 | -0.037 | -0.4371 | No | ||
| 123 | ATE1 | 290491 6940722 | 13849 | -0.038 | -0.4387 | No | ||
| 124 | PLCB3 | 4670402 | 13869 | -0.038 | -0.4387 | No | ||
| 125 | SYTL2 | 580097 5390576 6770603 | 14001 | -0.041 | -0.4448 | No | ||
| 126 | NRGN | 4280433 | 14020 | -0.041 | -0.4447 | No | ||
| 127 | A2BP1 | 2370390 4590593 5550014 | 14204 | -0.046 | -0.4535 | No | ||
| 128 | LPHN1 | 1400113 3130301 | 14414 | -0.051 | -0.4636 | No | ||
| 129 | NR1D1 | 2360471 770746 6590204 | 14471 | -0.053 | -0.4653 | No | ||
| 130 | IMPDH2 | 5220138 | 14487 | -0.053 | -0.4647 | No | ||
| 131 | TBCC | 4810021 | 14542 | -0.055 | -0.4663 | No | ||
| 132 | MAP1A | 4920576 | 14569 | -0.056 | -0.4663 | No | ||
| 133 | RPS10 | 4560673 | 14617 | -0.058 | -0.4674 | No | ||
| 134 | SLCO3A1 | 1050408 2370156 6110072 | 14744 | -0.061 | -0.4727 | No | ||
| 135 | DDR1 | 2060044 5220180 | 14951 | -0.069 | -0.4821 | No | ||
| 136 | PDGFB | 3060440 6370008 | 14969 | -0.070 | -0.4813 | No | ||
| 137 | TCF1 | 5390022 | 15045 | -0.073 | -0.4835 | No | ||
| 138 | PVRL1 | 2570167 5220519 5890500 3190309 | 15105 | -0.076 | -0.4847 | No | ||
| 139 | LBX1 | 1300164 4060204 | 15122 | -0.077 | -0.4837 | No | ||
| 140 | CLSTN3 | 2350685 | 15163 | -0.079 | -0.4839 | No | ||
| 141 | AP1G2 | 780458 2690068 | 15203 | -0.081 | -0.4839 | No | ||
| 142 | SYNPO2L | 5390719 | 15320 | -0.088 | -0.4880 | No | ||
| 143 | ITPR3 | 4010632 | 15594 | -0.105 | -0.5002 | No | ||
| 144 | RAB10 | 2360008 5890020 | 15646 | -0.108 | -0.5002 | No | ||
| 145 | SPI1 | 1410397 | 15838 | -0.125 | -0.5074 | No | ||
| 146 | DPYSL2 | 2100427 3130112 5700324 | 15925 | -0.133 | -0.5087 | No | ||
| 147 | TYRO3 | 3130193 | 16230 | -0.166 | -0.5210 | No | ||
| 148 | STAT6 | 1190010 5720019 | 16427 | -0.191 | -0.5269 | No | ||
| 149 | CLIC1 | 110600 | 16455 | -0.194 | -0.5234 | No | ||
| 150 | ARHGDIB | 4730433 | 16528 | -0.204 | -0.5222 | No | ||
| 151 | PEX16 | 4560044 | 16705 | -0.229 | -0.5260 | No | ||
| 152 | FOXP4 | 6290022 | 16742 | -0.235 | -0.5220 | No | ||
| 153 | DCTN1 | 1450056 6590022 | 16944 | -0.265 | -0.5263 | No | ||
| 154 | MRPS18B | 5570086 | 17340 | -0.340 | -0.5391 | No | ||
| 155 | GRWD1 | 6350528 | 17573 | -0.387 | -0.5419 | Yes | ||
| 156 | CDC42EP4 | 4150338 | 17605 | -0.397 | -0.5336 | Yes | ||
| 157 | MYL6 | 60563 6100152 | 17688 | -0.423 | -0.5274 | Yes | ||
| 158 | MTMR3 | 1050014 4920180 | 17763 | -0.450 | -0.5201 | Yes | ||
| 159 | RHEBL1 | 2810576 | 17796 | -0.460 | -0.5103 | Yes | ||
| 160 | CEBPB | 2970019 | 17856 | -0.485 | -0.5013 | Yes | ||
| 161 | ARFIP2 | 1850301 4670180 | 17865 | -0.489 | -0.4894 | Yes | ||
| 162 | RHOG | 6760575 | 17932 | -0.516 | -0.4800 | Yes | ||
| 163 | PTK2B | 4730411 | 18025 | -0.567 | -0.4707 | Yes | ||
| 164 | FXC1 | 3710053 4060575 | 18043 | -0.580 | -0.4570 | Yes | ||
| 165 | HSPG2 | 2510687 6220750 | 18163 | -0.669 | -0.4466 | Yes | ||
| 166 | DLK1 | 1240138 | 18381 | -0.977 | -0.4338 | Yes | ||
| 167 | SSBP2 | 2260725 2900400 4730711 | 18396 | -1.002 | -0.4093 | Yes | ||
| 168 | IMMT | 870450 4280438 | 18452 | -1.111 | -0.3844 | Yes | ||
| 169 | PDXP | 5050204 460273 6040102 | 18470 | -1.176 | -0.3557 | Yes | ||
| 170 | NDRG2 | 450403 | 18512 | -1.446 | -0.3215 | Yes | ||
| 171 | DTX1 | 5900372 | 18514 | -1.475 | -0.2844 | Yes | ||
| 172 | DNMT3B | 2320138 4070059 | 18522 | -1.518 | -0.2466 | Yes | ||
| 173 | CORO1A | 3140609 3190020 3190037 | 18548 | -1.660 | -0.2062 | Yes | ||
| 174 | ETV5 | 110017 | 18562 | -1.975 | -0.1571 | Yes | ||
| 175 | GNAS | 630441 1850373 4050152 | 18597 | -2.879 | -0.0865 | Yes | ||
| 176 | RHOA | 580142 5900131 5340450 | 18605 | -3.476 | 0.0006 | Yes |

