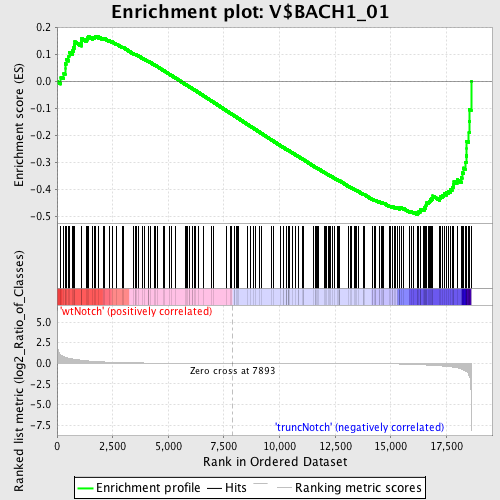
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$BACH1_01 |
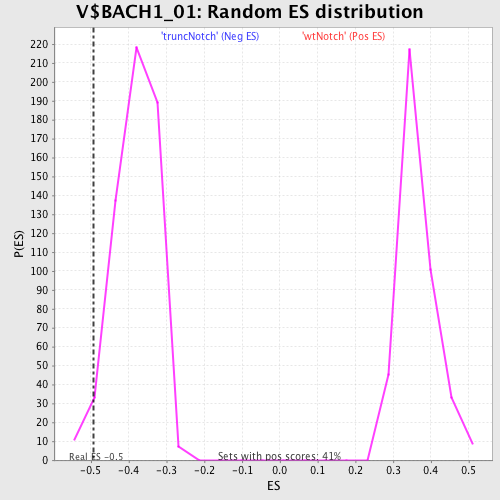
| Enrichment Score (ES) | -0.49377587 |
| Normalized Enrichment Score (NES) | -1.2945791 |
| Nominal p-value | 0.031932775 |
| FDR q-value | 0.82061833 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BRD2 | 1450300 5340484 6100605 | 172 | 0.960 | 0.0165 | No | ||
| 2 | SRPK2 | 6380341 | 304 | 0.780 | 0.0304 | No | ||
| 3 | H3F3B | 1410300 | 359 | 0.727 | 0.0470 | No | ||
| 4 | PPP2CA | 3990113 | 361 | 0.725 | 0.0665 | No | ||
| 5 | RTN4 | 50725 630647 870097 870184 | 402 | 0.685 | 0.0828 | No | ||
| 6 | LUC7L | 6040156 6110411 | 528 | 0.608 | 0.0924 | No | ||
| 7 | PSMD7 | 2030619 6220594 | 536 | 0.604 | 0.1083 | No | ||
| 8 | EEF1A1 | 1980193 1990195 4670735 | 693 | 0.517 | 0.1137 | No | ||
| 9 | TRIM8 | 6130441 | 733 | 0.496 | 0.1249 | No | ||
| 10 | ATP1B1 | 3130594 | 766 | 0.483 | 0.1362 | No | ||
| 11 | EIF4G1 | 4070446 | 800 | 0.472 | 0.1471 | No | ||
| 12 | PSMD12 | 730044 | 1085 | 0.365 | 0.1415 | No | ||
| 13 | RAB3D | 4210253 | 1097 | 0.359 | 0.1506 | No | ||
| 14 | RBBP7 | 430113 450450 2370309 | 1106 | 0.357 | 0.1598 | No | ||
| 15 | RAB34 | 770411 | 1330 | 0.299 | 0.1558 | No | ||
| 16 | MECP2 | 1940450 | 1363 | 0.290 | 0.1618 | No | ||
| 17 | LAMB3 | 7100301 | 1388 | 0.282 | 0.1681 | No | ||
| 18 | GTF2B | 6860685 | 1592 | 0.236 | 0.1634 | No | ||
| 19 | MPV17 | 1190133 4070427 4570577 | 1677 | 0.221 | 0.1648 | No | ||
| 20 | PSMD4 | 430068 | 1717 | 0.215 | 0.1685 | No | ||
| 21 | ABHD4 | 6760450 | 1856 | 0.188 | 0.1661 | No | ||
| 22 | APOBEC1 | 4610022 | 2070 | 0.157 | 0.1587 | No | ||
| 23 | ABI3 | 1740563 | 2107 | 0.151 | 0.1609 | No | ||
| 24 | ACADVL | 6040403 | 2332 | 0.123 | 0.1520 | No | ||
| 25 | GTPBP2 | 2760750 | 2476 | 0.108 | 0.1472 | No | ||
| 26 | BCL9L | 6940053 | 2690 | 0.090 | 0.1380 | No | ||
| 27 | DYRK1A | 3190181 | 2922 | 0.073 | 0.1274 | No | ||
| 28 | EN1 | 5360168 7050025 | 2979 | 0.069 | 0.1263 | No | ||
| 29 | LAMC1 | 4730025 | 3413 | 0.045 | 0.1040 | No | ||
| 30 | RPL23A | 1450403 | 3515 | 0.042 | 0.0996 | No | ||
| 31 | RIN1 | 510593 | 3517 | 0.041 | 0.1007 | No | ||
| 32 | NLK | 2030010 2450041 | 3545 | 0.040 | 0.1003 | No | ||
| 33 | TRAF3 | 5690647 | 3649 | 0.036 | 0.0957 | No | ||
| 34 | LAMA3 | 1190131 5290239 | 3826 | 0.031 | 0.0870 | No | ||
| 35 | TNRC15 | 6760746 | 3907 | 0.029 | 0.0834 | No | ||
| 36 | SSH2 | 360056 | 4096 | 0.024 | 0.0738 | No | ||
| 37 | HCN3 | 3940438 6840750 | 4112 | 0.024 | 0.0737 | No | ||
| 38 | RPS20 | 1740092 | 4127 | 0.024 | 0.0736 | No | ||
| 39 | ENO2 | 2320068 | 4193 | 0.023 | 0.0706 | No | ||
| 40 | CAPN12 | 2100050 | 4377 | 0.020 | 0.0612 | No | ||
| 41 | TNXB | 630592 3830020 5360497 7000673 | 4440 | 0.019 | 0.0584 | No | ||
| 42 | LRRC15 | 3940609 | 4524 | 0.018 | 0.0544 | No | ||
| 43 | SPARC | 1690086 | 4792 | 0.015 | 0.0403 | No | ||
| 44 | TIAL1 | 4150048 6510605 | 4834 | 0.014 | 0.0384 | No | ||
| 45 | PSMD11 | 2340538 6510053 | 5043 | 0.012 | 0.0275 | No | ||
| 46 | CDKN1A | 4050088 6400706 | 5145 | 0.011 | 0.0223 | No | ||
| 47 | KCNA2 | 5080403 | 5325 | 0.010 | 0.0128 | No | ||
| 48 | TAC1 | 7000195 380706 | 5783 | 0.007 | -0.0118 | No | ||
| 49 | P2RXL1 | 4730390 5220446 | 5798 | 0.007 | -0.0123 | No | ||
| 50 | UCN2 | 4590372 | 5879 | 0.007 | -0.0165 | No | ||
| 51 | SNCB | 3360133 | 5942 | 0.007 | -0.0197 | No | ||
| 52 | GRIA1 | 1340152 3780750 4920440 | 5971 | 0.007 | -0.0210 | No | ||
| 53 | MYH14 | 4010017 | 6066 | 0.006 | -0.0259 | No | ||
| 54 | GFAP | 2060092 | 6098 | 0.006 | -0.0275 | No | ||
| 55 | IL11 | 1740398 | 6195 | 0.006 | -0.0325 | No | ||
| 56 | ADAMTSL1 | 2970092 | 6204 | 0.005 | -0.0328 | No | ||
| 57 | GNAI1 | 4560390 | 6364 | 0.005 | -0.0413 | No | ||
| 58 | FAP | 4560019 | 6584 | 0.004 | -0.0531 | No | ||
| 59 | FGF12 | 1740446 2360037 | 6946 | 0.003 | -0.0726 | No | ||
| 60 | AKAP1 | 110148 1740735 2260019 7000563 | 7026 | 0.003 | -0.0768 | No | ||
| 61 | NR1I3 | 50026 1240673 | 7595 | 0.001 | -0.1076 | No | ||
| 62 | RGS2 | 1090736 | 7779 | 0.000 | -0.1175 | No | ||
| 63 | SLC6A5 | 870121 | 7826 | 0.000 | -0.1200 | No | ||
| 64 | AKT3 | 1580270 3290278 | 7974 | -0.000 | -0.1280 | No | ||
| 65 | IL1RN | 2370333 | 7976 | -0.000 | -0.1280 | No | ||
| 66 | MMP3 | 2650368 2970056 | 8079 | -0.001 | -0.1336 | No | ||
| 67 | SLC24A4 | 2100487 | 8090 | -0.001 | -0.1341 | No | ||
| 68 | SYT2 | 60609 | 8157 | -0.001 | -0.1377 | No | ||
| 69 | PHLDA2 | 4810494 | 8570 | -0.002 | -0.1600 | No | ||
| 70 | FABP4 | 5050114 | 8675 | -0.002 | -0.1655 | No | ||
| 71 | MYB | 1660494 5860451 6130706 | 8839 | -0.003 | -0.1743 | No | ||
| 72 | GPR56 | 1190494 | 8917 | -0.003 | -0.1784 | No | ||
| 73 | LY6G6C | 6020673 | 9081 | -0.003 | -0.1872 | No | ||
| 74 | SYP | 2690168 | 9188 | -0.004 | -0.1928 | No | ||
| 75 | SH3RF2 | 6200600 | 9656 | -0.005 | -0.2180 | No | ||
| 76 | EIF3S1 | 6130368 6770044 | 9725 | -0.005 | -0.2216 | No | ||
| 77 | OSBP | 5390471 | 10037 | -0.006 | -0.2383 | No | ||
| 78 | IBRDC2 | 5130041 | 10160 | -0.007 | -0.2447 | No | ||
| 79 | MAMDC2 | 430471 | 10169 | -0.007 | -0.2450 | No | ||
| 80 | LRRTM3 | 4210021 | 10295 | -0.007 | -0.2516 | No | ||
| 81 | BMP2 | 6620687 | 10310 | -0.007 | -0.2521 | No | ||
| 82 | BNIP3 | 3140270 | 10311 | -0.007 | -0.2519 | No | ||
| 83 | TLL1 | 2680315 | 10397 | -0.008 | -0.2563 | No | ||
| 84 | NTN4 | 6940398 | 10399 | -0.008 | -0.2562 | No | ||
| 85 | DSC2 | 840484 | 10414 | -0.008 | -0.2568 | No | ||
| 86 | LPP | 1660541 | 10447 | -0.008 | -0.2583 | No | ||
| 87 | MYBPH | 2190711 | 10575 | -0.008 | -0.2649 | No | ||
| 88 | SLC26A1 | 2190142 | 10698 | -0.009 | -0.2713 | No | ||
| 89 | CAPN6 | 1740168 | 10843 | -0.009 | -0.2789 | No | ||
| 90 | FOXA1 | 3710609 | 10870 | -0.009 | -0.2800 | No | ||
| 91 | GDNF | 4050487 5720739 | 11018 | -0.010 | -0.2877 | No | ||
| 92 | SHC3 | 2690398 | 11057 | -0.010 | -0.2895 | No | ||
| 93 | FADS3 | 3120079 | 11063 | -0.010 | -0.2895 | No | ||
| 94 | TGM3 | 840576 | 11515 | -0.013 | -0.3137 | No | ||
| 95 | HOXB3 | 520292 | 11609 | -0.013 | -0.3184 | No | ||
| 96 | BTK | 3130044 | 11612 | -0.013 | -0.3181 | No | ||
| 97 | PADI3 | 6520072 | 11660 | -0.013 | -0.3203 | No | ||
| 98 | DIRAS1 | 2640270 | 11693 | -0.014 | -0.3217 | No | ||
| 99 | PPP1R9B | 3130619 | 11732 | -0.014 | -0.3234 | No | ||
| 100 | ELK3 | 460377 1400762 3710497 | 12000 | -0.016 | -0.3374 | No | ||
| 101 | VIT | 6620301 | 12085 | -0.016 | -0.3415 | No | ||
| 102 | HS3ST3B1 | 380040 | 12112 | -0.016 | -0.3425 | No | ||
| 103 | PTPRR | 130121 3990273 | 12200 | -0.017 | -0.3468 | No | ||
| 104 | DNAJC5B | 60368 | 12249 | -0.018 | -0.3489 | No | ||
| 105 | TRIB1 | 2320435 | 12289 | -0.018 | -0.3505 | No | ||
| 106 | NEK6 | 3360687 | 12367 | -0.019 | -0.3542 | No | ||
| 107 | FOXN1 | 4730075 | 12482 | -0.020 | -0.3599 | No | ||
| 108 | MARK1 | 450484 | 12605 | -0.021 | -0.3659 | No | ||
| 109 | GKN1 | 5420091 | 12630 | -0.021 | -0.3667 | No | ||
| 110 | ELAVL2 | 360181 | 12702 | -0.022 | -0.3699 | No | ||
| 111 | EEF1A2 | 2260162 5290086 | 12703 | -0.022 | -0.3694 | No | ||
| 112 | LAMC2 | 450692 2680041 4200059 4670148 | 12714 | -0.022 | -0.3693 | No | ||
| 113 | DYSF | 4670411 | 13086 | -0.026 | -0.3888 | No | ||
| 114 | GPR3 | 2970452 | 13186 | -0.027 | -0.3934 | No | ||
| 115 | F2RL2 | 6110736 | 13236 | -0.028 | -0.3953 | No | ||
| 116 | PROK2 | 540538 | 13358 | -0.030 | -0.4011 | No | ||
| 117 | CSF3 | 2230193 6660707 | 13421 | -0.031 | -0.4036 | No | ||
| 118 | UBE2C | 6130017 | 13458 | -0.031 | -0.4047 | No | ||
| 119 | TREX1 | 3450040 7100692 | 13559 | -0.033 | -0.4093 | No | ||
| 120 | SYNGR1 | 5130452 6650056 | 13763 | -0.036 | -0.4193 | No | ||
| 121 | PKP3 | 3710048 | 13780 | -0.037 | -0.4192 | No | ||
| 122 | SYN1 | 4920301 7050440 | 13822 | -0.037 | -0.4204 | No | ||
| 123 | MRGPRF | 4780746 | 14154 | -0.045 | -0.4371 | No | ||
| 124 | RB1CC1 | 510494 7100072 | 14171 | -0.045 | -0.4368 | No | ||
| 125 | PLEC1 | 6520292 50500 380075 1570088 2510692 2630497 2680017 2900746 4560497 4850376 6290398 7040471 | 14267 | -0.047 | -0.4407 | No | ||
| 126 | CSMD3 | 1660427 1940687 | 14291 | -0.047 | -0.4407 | No | ||
| 127 | TRIM47 | 5080717 | 14305 | -0.048 | -0.4401 | No | ||
| 128 | NR1D1 | 2360471 770746 6590204 | 14471 | -0.053 | -0.4476 | No | ||
| 129 | NRAS | 6900577 | 14481 | -0.053 | -0.4467 | No | ||
| 130 | DCTN2 | 540471 3780717 | 14598 | -0.057 | -0.4514 | No | ||
| 131 | PRX | 2260400 4200347 | 14606 | -0.057 | -0.4503 | No | ||
| 132 | DDIT3 | 780373 | 14666 | -0.059 | -0.4519 | No | ||
| 133 | COL7A1 | 1230168 2190072 | 14692 | -0.060 | -0.4516 | No | ||
| 134 | EIF4E | 1580403 70133 6380215 | 14939 | -0.069 | -0.4631 | No | ||
| 135 | SERTAD1 | 2680332 | 14981 | -0.071 | -0.4634 | No | ||
| 136 | ACPP | 2470152 1780593 2690524 | 15069 | -0.075 | -0.4661 | No | ||
| 137 | BDKRB1 | 730592 | 15075 | -0.075 | -0.4644 | No | ||
| 138 | ELL | 3520731 | 15177 | -0.080 | -0.4677 | No | ||
| 139 | SLC26A9 | 3440722 5340088 7510093 | 15204 | -0.081 | -0.4670 | No | ||
| 140 | ADRA1A | 1230446 2260390 | 15314 | -0.088 | -0.4705 | No | ||
| 141 | CDH23 | 4250242 5290397 6550397 | 15379 | -0.091 | -0.4715 | No | ||
| 142 | CFL2 | 380239 6200368 | 15401 | -0.093 | -0.4702 | No | ||
| 143 | ITGB4 | 1740021 3840482 | 15402 | -0.093 | -0.4677 | No | ||
| 144 | WASF2 | 4850592 | 15474 | -0.098 | -0.4689 | No | ||
| 145 | SEC24D | 3060279 | 15575 | -0.104 | -0.4715 | No | ||
| 146 | PSME4 | 380348 1770131 | 15857 | -0.127 | -0.4833 | No | ||
| 147 | TUBB4 | 5310681 | 15921 | -0.132 | -0.4832 | No | ||
| 148 | PIM1 | 630047 | 16020 | -0.143 | -0.4847 | No | ||
| 149 | HDAC3 | 4060072 | 16189 | -0.161 | -0.4894 | Yes | ||
| 150 | AXUD1 | 1230053 | 16213 | -0.164 | -0.4863 | Yes | ||
| 151 | CAMKK1 | 6370324 | 16259 | -0.169 | -0.4842 | Yes | ||
| 152 | SCRN1 | 6040025 6580019 | 16314 | -0.177 | -0.4823 | Yes | ||
| 153 | ABCA2 | 5050039 | 16320 | -0.177 | -0.4778 | Yes | ||
| 154 | LTB4R2 | 1990746 | 16331 | -0.179 | -0.4736 | Yes | ||
| 155 | TNFRSF12A | 4810253 | 16448 | -0.193 | -0.4747 | Yes | ||
| 156 | MNT | 4920575 | 16500 | -0.200 | -0.4721 | Yes | ||
| 157 | ABCD1 | 5670373 | 16525 | -0.203 | -0.4679 | Yes | ||
| 158 | EMP3 | 1500722 1570039 | 16574 | -0.210 | -0.4648 | Yes | ||
| 159 | RPS6KA4 | 2190524 | 16579 | -0.211 | -0.4594 | Yes | ||
| 160 | GPX1 | 4150093 | 16587 | -0.212 | -0.4540 | Yes | ||
| 161 | EDG5 | 2640300 | 16621 | -0.218 | -0.4500 | Yes | ||
| 162 | ROM1 | 3130685 | 16695 | -0.228 | -0.4478 | Yes | ||
| 163 | SNPH | 2680484 | 16740 | -0.234 | -0.4439 | Yes | ||
| 164 | HSPB8 | 540563 | 16768 | -0.238 | -0.4390 | Yes | ||
| 165 | ADCK4 | 1660180 | 16821 | -0.247 | -0.4351 | Yes | ||
| 166 | AP2A2 | 130082 | 16891 | -0.257 | -0.4319 | Yes | ||
| 167 | TRAPPC3 | 2030176 6020609 6590373 6760019 | 16892 | -0.257 | -0.4250 | Yes | ||
| 168 | MAST2 | 6200348 | 17192 | -0.308 | -0.4329 | Yes | ||
| 169 | BAG2 | 2900324 | 17243 | -0.320 | -0.4270 | Yes | ||
| 170 | GABARAPL1 | 2810458 | 17321 | -0.336 | -0.4222 | Yes | ||
| 171 | HMGA1 | 6580408 | 17390 | -0.349 | -0.4165 | Yes | ||
| 172 | ITPKC | 5420021 | 17488 | -0.366 | -0.4119 | Yes | ||
| 173 | SQSTM1 | 6550056 | 17591 | -0.393 | -0.4068 | Yes | ||
| 174 | SGK | 1400131 2480056 | 17670 | -0.418 | -0.3998 | Yes | ||
| 175 | GNGT2 | 540176 | 17754 | -0.447 | -0.3923 | Yes | ||
| 176 | ALS2CR2 | 6450128 7100092 | 17807 | -0.464 | -0.3826 | Yes | ||
| 177 | ALDOA | 6290672 | 17837 | -0.478 | -0.3713 | Yes | ||
| 178 | TAGLN2 | 4560121 | 17977 | -0.534 | -0.3645 | Yes | ||
| 179 | LMNA | 520471 1500075 3190167 4210020 | 18166 | -0.670 | -0.3567 | Yes | ||
| 180 | S100A10 | 6350010 | 18202 | -0.712 | -0.3394 | Yes | ||
| 181 | ECM1 | 940068 5390309 | 18264 | -0.800 | -0.3211 | Yes | ||
| 182 | UBE2E3 | 2030347 3840471 | 18347 | -0.933 | -0.3005 | Yes | ||
| 183 | CHST1 | 50601 | 18387 | -0.989 | -0.2760 | Yes | ||
| 184 | EPHB2 | 1090288 1410377 2100541 | 18409 | -1.036 | -0.2492 | Yes | ||
| 185 | PADI4 | 1740075 4810441 | 18412 | -1.039 | -0.2213 | Yes | ||
| 186 | NDRG2 | 450403 | 18512 | -1.446 | -0.1878 | Yes | ||
| 187 | ENO1 | 5340128 | 18526 | -1.526 | -0.1474 | Yes | ||
| 188 | NCDN | 50040 1980739 3520603 | 18539 | -1.599 | -0.1050 | Yes | ||
| 189 | SFRS2 | 50707 380593 | 18609 | -4.052 | 0.0004 | Yes |