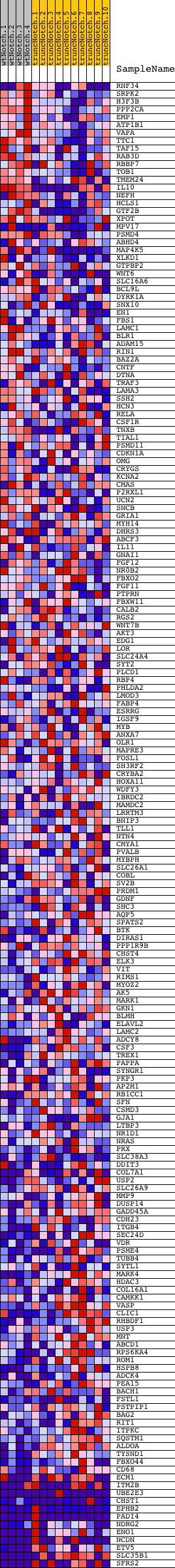
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
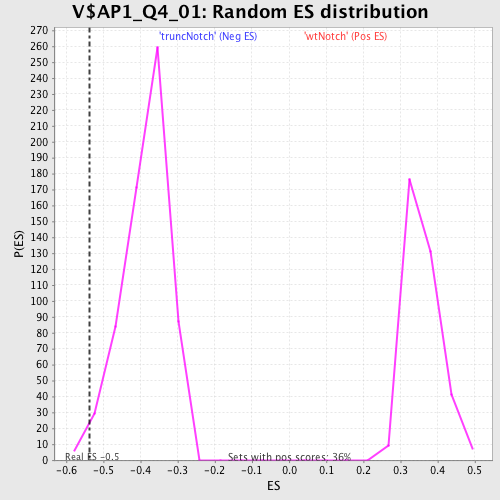
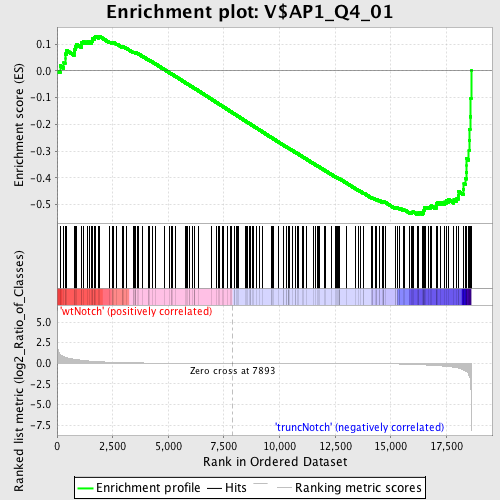
| GeneSet | V$AP1_Q4_01 |
| Enrichment Score (ES) | -0.53698134 |
| Normalized Enrichment Score (NES) | -1.392007 |
| Nominal p-value | 0.012578616 |
| FDR q-value | 0.8120526 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RNF34 | 3800332 4590717 5670138 | 137 | 1.062 | 0.0194 | No | ||
| 2 | SRPK2 | 6380341 | 304 | 0.780 | 0.0301 | No | ||
| 3 | H3F3B | 1410300 | 359 | 0.727 | 0.0455 | No | ||
| 4 | PPP2CA | 3990113 | 361 | 0.725 | 0.0638 | No | ||
| 5 | EMP1 | 4120438 | 432 | 0.662 | 0.0767 | No | ||
| 6 | ATP1B1 | 3130594 | 766 | 0.483 | 0.0709 | No | ||
| 7 | VAPA | 2480594 | 781 | 0.478 | 0.0822 | No | ||
| 8 | TTC1 | 5890736 | 827 | 0.459 | 0.0913 | No | ||
| 9 | TAF15 | 4120736 | 859 | 0.447 | 0.1010 | No | ||
| 10 | RAB3D | 4210253 | 1097 | 0.359 | 0.0972 | No | ||
| 11 | RBBP7 | 430113 450450 2370309 | 1106 | 0.357 | 0.1058 | No | ||
| 12 | TOB1 | 4150138 | 1181 | 0.340 | 0.1103 | No | ||
| 13 | TMEM24 | 5860039 | 1346 | 0.295 | 0.1089 | No | ||
| 14 | IL10 | 2340685 2640541 2850403 6590286 | 1468 | 0.263 | 0.1090 | No | ||
| 15 | NEFH | 630239 | 1565 | 0.242 | 0.1099 | No | ||
| 16 | HCLS1 | 7100309 | 1586 | 0.237 | 0.1148 | No | ||
| 17 | GTF2B | 6860685 | 1592 | 0.236 | 0.1205 | No | ||
| 18 | XPOT | 7050184 | 1670 | 0.222 | 0.1219 | No | ||
| 19 | MPV17 | 1190133 4070427 4570577 | 1677 | 0.221 | 0.1272 | No | ||
| 20 | PSMD4 | 430068 | 1717 | 0.215 | 0.1305 | No | ||
| 21 | ABHD4 | 6760450 | 1856 | 0.188 | 0.1278 | No | ||
| 22 | MAP4K5 | 3190286 4760300 5270070 5550494 | 1892 | 0.183 | 0.1305 | No | ||
| 23 | XLKD1 | 520441 | 2361 | 0.120 | 0.1081 | No | ||
| 24 | GTPBP2 | 2760750 | 2476 | 0.108 | 0.1046 | No | ||
| 25 | WNT6 | 4570364 | 2497 | 0.106 | 0.1062 | No | ||
| 26 | SLC16A6 | 1690156 | 2524 | 0.104 | 0.1074 | No | ||
| 27 | BCL9L | 6940053 | 2690 | 0.090 | 0.1008 | No | ||
| 28 | DYRK1A | 3190181 | 2922 | 0.073 | 0.0901 | No | ||
| 29 | SNX10 | 3800035 | 2939 | 0.072 | 0.0910 | No | ||
| 30 | EN1 | 5360168 7050025 | 2979 | 0.069 | 0.0906 | No | ||
| 31 | FBS1 | 2570520 | 3129 | 0.060 | 0.0841 | No | ||
| 32 | LAMC1 | 4730025 | 3413 | 0.045 | 0.0698 | No | ||
| 33 | BLR1 | 1500300 5420017 | 3445 | 0.044 | 0.0693 | No | ||
| 34 | ADAM15 | 5890707 | 3459 | 0.044 | 0.0697 | No | ||
| 35 | RIN1 | 510593 | 3517 | 0.041 | 0.0676 | No | ||
| 36 | BAZ2A | 730184 | 3525 | 0.041 | 0.0683 | No | ||
| 37 | CNTF | 450128 870253 1780068 6520195 | 3526 | 0.041 | 0.0693 | No | ||
| 38 | DTNA | 1340600 1780731 2340278 2850132 | 3621 | 0.037 | 0.0651 | No | ||
| 39 | TRAF3 | 5690647 | 3649 | 0.036 | 0.0646 | No | ||
| 40 | LAMA3 | 1190131 5290239 | 3826 | 0.031 | 0.0558 | No | ||
| 41 | SSH2 | 360056 | 4096 | 0.024 | 0.0418 | No | ||
| 42 | HCN3 | 3940438 6840750 | 4112 | 0.024 | 0.0416 | No | ||
| 43 | RELA | 3830075 | 4139 | 0.024 | 0.0408 | No | ||
| 44 | CSF1R | 2340110 6420408 | 4292 | 0.021 | 0.0331 | No | ||
| 45 | TNXB | 630592 3830020 5360497 7000673 | 4440 | 0.019 | 0.0256 | No | ||
| 46 | TIAL1 | 4150048 6510605 | 4834 | 0.014 | 0.0046 | No | ||
| 47 | PSMD11 | 2340538 6510053 | 5043 | 0.012 | -0.0064 | No | ||
| 48 | CDKN1A | 4050088 6400706 | 5145 | 0.011 | -0.0115 | No | ||
| 49 | OMG | 6760066 | 5146 | 0.011 | -0.0113 | No | ||
| 50 | CRYGS | 2680398 | 5192 | 0.011 | -0.0134 | No | ||
| 51 | KCNA2 | 5080403 | 5325 | 0.010 | -0.0203 | No | ||
| 52 | CMAS | 2190129 | 5759 | 0.008 | -0.0436 | No | ||
| 53 | P2RXL1 | 4730390 5220446 | 5798 | 0.007 | -0.0455 | No | ||
| 54 | UCN2 | 4590372 | 5879 | 0.007 | -0.0497 | No | ||
| 55 | SNCB | 3360133 | 5942 | 0.007 | -0.0529 | No | ||
| 56 | GRIA1 | 1340152 3780750 4920440 | 5971 | 0.007 | -0.0542 | No | ||
| 57 | MYH14 | 4010017 | 6066 | 0.006 | -0.0592 | No | ||
| 58 | DHRS3 | 360609 | 6184 | 0.006 | -0.0654 | No | ||
| 59 | ABCF3 | 7100100 | 6191 | 0.006 | -0.0656 | No | ||
| 60 | IL11 | 1740398 | 6195 | 0.006 | -0.0656 | No | ||
| 61 | GNAI1 | 4560390 | 6364 | 0.005 | -0.0746 | No | ||
| 62 | FGF12 | 1740446 2360037 | 6946 | 0.003 | -0.1060 | No | ||
| 63 | NR0B2 | 2470692 | 7141 | 0.002 | -0.1165 | No | ||
| 64 | FBXO2 | 6510091 | 7248 | 0.002 | -0.1222 | No | ||
| 65 | FGF11 | 840292 | 7297 | 0.002 | -0.1248 | No | ||
| 66 | PTPRN | 5900577 | 7449 | 0.001 | -0.1330 | No | ||
| 67 | FBXW11 | 6450632 | 7459 | 0.001 | -0.1334 | No | ||
| 68 | CALB2 | 6130170 6590368 | 7665 | 0.001 | -0.1445 | No | ||
| 69 | RGS2 | 1090736 | 7779 | 0.000 | -0.1506 | No | ||
| 70 | WNT7B | 3520039 | 7818 | 0.000 | -0.1527 | No | ||
| 71 | AKT3 | 1580270 3290278 | 7974 | -0.000 | -0.1611 | No | ||
| 72 | EDG1 | 4200619 | 7994 | -0.000 | -0.1621 | No | ||
| 73 | LOR | 50538 | 8062 | -0.000 | -0.1658 | No | ||
| 74 | SLC24A4 | 2100487 | 8090 | -0.001 | -0.1672 | No | ||
| 75 | SYT2 | 60609 | 8157 | -0.001 | -0.1708 | No | ||
| 76 | PLCD1 | 2030484 | 8480 | -0.002 | -0.1882 | No | ||
| 77 | RBP4 | 4540131 | 8528 | -0.002 | -0.1907 | No | ||
| 78 | PHLDA2 | 4810494 | 8570 | -0.002 | -0.1929 | No | ||
| 79 | LMOD3 | 2120524 | 8641 | -0.002 | -0.1966 | No | ||
| 80 | FABP4 | 5050114 | 8675 | -0.002 | -0.1984 | No | ||
| 81 | ESRRG | 4010181 | 8681 | -0.002 | -0.1986 | No | ||
| 82 | IGSF9 | 1770411 | 8763 | -0.002 | -0.2029 | No | ||
| 83 | MYB | 1660494 5860451 6130706 | 8839 | -0.003 | -0.2069 | No | ||
| 84 | ANXA7 | 6110446 | 8963 | -0.003 | -0.2135 | No | ||
| 85 | OLR1 | 50025 3840484 | 9104 | -0.003 | -0.2210 | No | ||
| 86 | MAPRE3 | 7050504 | 9213 | -0.004 | -0.2268 | No | ||
| 87 | FOSL1 | 430021 | 9620 | -0.005 | -0.2487 | No | ||
| 88 | SH3RF2 | 6200600 | 9656 | -0.005 | -0.2505 | No | ||
| 89 | CRYBA2 | 5900138 | 9667 | -0.005 | -0.2509 | No | ||
| 90 | HOXA11 | 6980133 | 9741 | -0.005 | -0.2547 | No | ||
| 91 | WDFY3 | 3610041 4560333 4570273 | 9963 | -0.006 | -0.2666 | No | ||
| 92 | IBRDC2 | 5130041 | 10160 | -0.007 | -0.2770 | No | ||
| 93 | MAMDC2 | 430471 | 10169 | -0.007 | -0.2773 | No | ||
| 94 | LRRTM3 | 4210021 | 10295 | -0.007 | -0.2839 | No | ||
| 95 | BNIP3 | 3140270 | 10311 | -0.007 | -0.2845 | No | ||
| 96 | TLL1 | 2680315 | 10397 | -0.008 | -0.2889 | No | ||
| 97 | NTN4 | 6940398 | 10399 | -0.008 | -0.2888 | No | ||
| 98 | CMYA1 | 6900632 | 10404 | -0.008 | -0.2888 | No | ||
| 99 | PVALB | 5570450 | 10449 | -0.008 | -0.2910 | No | ||
| 100 | MYBPH | 2190711 | 10575 | -0.008 | -0.2976 | No | ||
| 101 | SLC26A1 | 2190142 | 10698 | -0.009 | -0.3040 | No | ||
| 102 | COBL | 2480452 6110066 | 10700 | -0.009 | -0.3038 | No | ||
| 103 | SV2B | 3850717 | 10799 | -0.009 | -0.3089 | No | ||
| 104 | PRDM1 | 3170347 3520301 | 10835 | -0.009 | -0.3106 | No | ||
| 105 | GDNF | 4050487 5720739 | 11018 | -0.010 | -0.3202 | No | ||
| 106 | SHC3 | 2690398 | 11057 | -0.010 | -0.3220 | No | ||
| 107 | AQP5 | 3830451 | 11217 | -0.011 | -0.3303 | No | ||
| 108 | SPATS2 | 2360066 | 11506 | -0.013 | -0.3456 | No | ||
| 109 | BTK | 3130044 | 11612 | -0.013 | -0.3510 | No | ||
| 110 | DIRAS1 | 2640270 | 11693 | -0.014 | -0.3550 | No | ||
| 111 | PPP1R9B | 3130619 | 11732 | -0.014 | -0.3567 | No | ||
| 112 | CHST4 | 1190008 | 11791 | -0.014 | -0.3595 | No | ||
| 113 | ELK3 | 460377 1400762 3710497 | 12000 | -0.016 | -0.3704 | No | ||
| 114 | VIT | 6620301 | 12085 | -0.016 | -0.3745 | No | ||
| 115 | RIMS1 | 5670022 | 12325 | -0.018 | -0.3871 | No | ||
| 116 | MYOZ2 | 1050253 | 12503 | -0.020 | -0.3962 | No | ||
| 117 | AK5 | 6290348 | 12555 | -0.020 | -0.3984 | No | ||
| 118 | MARK1 | 450484 | 12605 | -0.021 | -0.4006 | No | ||
| 119 | GKN1 | 5420091 | 12630 | -0.021 | -0.4013 | No | ||
| 120 | BLMH | 1410438 | 12677 | -0.021 | -0.4033 | No | ||
| 121 | ELAVL2 | 360181 | 12702 | -0.022 | -0.4040 | No | ||
| 122 | LAMC2 | 450692 2680041 4200059 4670148 | 12714 | -0.022 | -0.4041 | No | ||
| 123 | ADCY8 | 6760519 | 12989 | -0.025 | -0.4183 | No | ||
| 124 | CSF3 | 2230193 6660707 | 13421 | -0.031 | -0.4409 | No | ||
| 125 | TREX1 | 3450040 7100692 | 13559 | -0.033 | -0.4475 | No | ||
| 126 | PAPPA | 4230463 | 13641 | -0.034 | -0.4511 | No | ||
| 127 | SYNGR1 | 5130452 6650056 | 13763 | -0.036 | -0.4567 | No | ||
| 128 | PKP3 | 3710048 | 13780 | -0.037 | -0.4567 | No | ||
| 129 | AP2M1 | 2570338 | 14121 | -0.044 | -0.4740 | No | ||
| 130 | RB1CC1 | 510494 7100072 | 14171 | -0.045 | -0.4755 | No | ||
| 131 | SFN | 6290301 7510608 | 14188 | -0.045 | -0.4752 | No | ||
| 132 | CSMD3 | 1660427 1940687 | 14291 | -0.047 | -0.4796 | No | ||
| 133 | GJA1 | 5220731 | 14358 | -0.049 | -0.4819 | No | ||
| 134 | LTBP3 | 1190162 2650440 6520451 | 14374 | -0.050 | -0.4815 | No | ||
| 135 | NR1D1 | 2360471 770746 6590204 | 14471 | -0.053 | -0.4853 | No | ||
| 136 | NRAS | 6900577 | 14481 | -0.053 | -0.4845 | No | ||
| 137 | PRX | 2260400 4200347 | 14606 | -0.057 | -0.4898 | No | ||
| 138 | SLC38A3 | 5340142 | 14653 | -0.059 | -0.4908 | No | ||
| 139 | DDIT3 | 780373 | 14666 | -0.059 | -0.4899 | No | ||
| 140 | COL7A1 | 1230168 2190072 | 14692 | -0.060 | -0.4898 | No | ||
| 141 | USP2 | 1190292 1240253 4850035 | 14758 | -0.062 | -0.4918 | No | ||
| 142 | SLC26A9 | 3440722 5340088 7510093 | 15204 | -0.081 | -0.5139 | No | ||
| 143 | MMP9 | 580338 | 15215 | -0.082 | -0.5123 | No | ||
| 144 | DUSP14 | 1230358 | 15291 | -0.086 | -0.5142 | No | ||
| 145 | GADD45A | 2900717 | 15292 | -0.086 | -0.5120 | No | ||
| 146 | CDH23 | 4250242 5290397 6550397 | 15379 | -0.091 | -0.5144 | No | ||
| 147 | ITGB4 | 1740021 3840482 | 15402 | -0.093 | -0.5132 | No | ||
| 148 | SEC24D | 3060279 | 15575 | -0.104 | -0.5200 | No | ||
| 149 | VDR | 130156 510438 | 15628 | -0.107 | -0.5201 | No | ||
| 150 | PSME4 | 380348 1770131 | 15857 | -0.127 | -0.5292 | No | ||
| 151 | TUBB4 | 5310681 | 15921 | -0.132 | -0.5293 | No | ||
| 152 | SYTL1 | 510487 | 15953 | -0.136 | -0.5276 | No | ||
| 153 | MARK4 | 730519 | 16015 | -0.142 | -0.5273 | No | ||
| 154 | HDAC3 | 4060072 | 16189 | -0.161 | -0.5326 | Yes | ||
| 155 | COL16A1 | 1780520 | 16247 | -0.168 | -0.5315 | Yes | ||
| 156 | CAMKK1 | 6370324 | 16259 | -0.169 | -0.5278 | Yes | ||
| 157 | VASP | 7050500 | 16429 | -0.191 | -0.5322 | Yes | ||
| 158 | CLIC1 | 110600 | 16455 | -0.194 | -0.5286 | Yes | ||
| 159 | RHBDF1 | 380538 | 16456 | -0.194 | -0.5237 | Yes | ||
| 160 | USP3 | 2060332 | 16490 | -0.198 | -0.5205 | Yes | ||
| 161 | MNT | 4920575 | 16500 | -0.200 | -0.5159 | Yes | ||
| 162 | ABCD1 | 5670373 | 16525 | -0.203 | -0.5121 | Yes | ||
| 163 | RPS6KA4 | 2190524 | 16579 | -0.211 | -0.5096 | Yes | ||
| 164 | ROM1 | 3130685 | 16695 | -0.228 | -0.5101 | Yes | ||
| 165 | HSPB8 | 540563 | 16768 | -0.238 | -0.5080 | Yes | ||
| 166 | ADCK4 | 1660180 | 16821 | -0.247 | -0.5046 | Yes | ||
| 167 | PEA15 | 4590685 | 17036 | -0.281 | -0.5091 | Yes | ||
| 168 | BACH1 | 290195 | 17038 | -0.281 | -0.5021 | Yes | ||
| 169 | FSTL1 | 2900113 | 17046 | -0.282 | -0.4953 | Yes | ||
| 170 | PSTPIP1 | 3190156 | 17111 | -0.293 | -0.4914 | Yes | ||
| 171 | BAG2 | 2900324 | 17243 | -0.320 | -0.4904 | Yes | ||
| 172 | RIT1 | 5390338 | 17404 | -0.352 | -0.4902 | Yes | ||
| 173 | ITPKC | 5420021 | 17488 | -0.366 | -0.4855 | Yes | ||
| 174 | SQSTM1 | 6550056 | 17591 | -0.393 | -0.4811 | Yes | ||
| 175 | ALDOA | 6290672 | 17837 | -0.478 | -0.4823 | Yes | ||
| 176 | TYSND1 | 2940273 | 17968 | -0.532 | -0.4759 | Yes | ||
| 177 | FBXO44 | 4760372 | 18031 | -0.570 | -0.4648 | Yes | ||
| 178 | CD68 | 6350450 | 18041 | -0.578 | -0.4507 | Yes | ||
| 179 | ECM1 | 940068 5390309 | 18264 | -0.800 | -0.4426 | Yes | ||
| 180 | ITM2B | 1850482 5860671 6200014 | 18287 | -0.839 | -0.4225 | Yes | ||
| 181 | UBE2E3 | 2030347 3840471 | 18347 | -0.933 | -0.4022 | Yes | ||
| 182 | CHST1 | 50601 | 18387 | -0.989 | -0.3793 | Yes | ||
| 183 | EPHB2 | 1090288 1410377 2100541 | 18409 | -1.036 | -0.3543 | Yes | ||
| 184 | PADI4 | 1740075 4810441 | 18412 | -1.039 | -0.3281 | Yes | ||
| 185 | NDRG2 | 450403 | 18512 | -1.446 | -0.2969 | Yes | ||
| 186 | ENO1 | 5340128 | 18526 | -1.526 | -0.2591 | Yes | ||
| 187 | NCDN | 50040 1980739 3520603 | 18539 | -1.599 | -0.2193 | Yes | ||
| 188 | ETV5 | 110017 | 18562 | -1.975 | -0.1706 | Yes | ||
| 189 | SLC35B1 | 5700064 7000735 | 18596 | -2.812 | -0.1013 | Yes | ||
| 190 | SFRS2 | 50707 380593 | 18609 | -4.052 | 0.0004 | Yes |