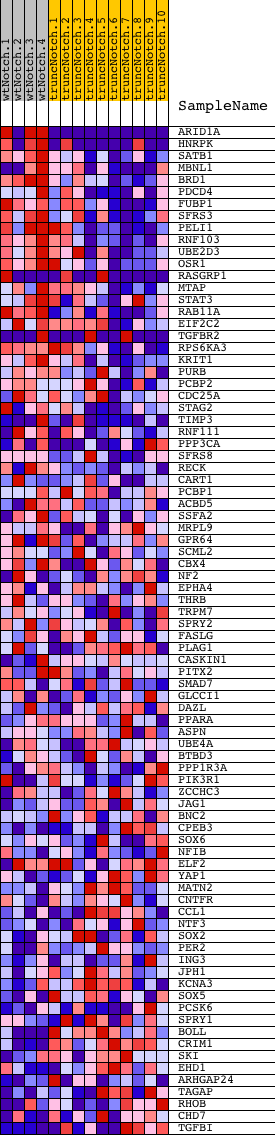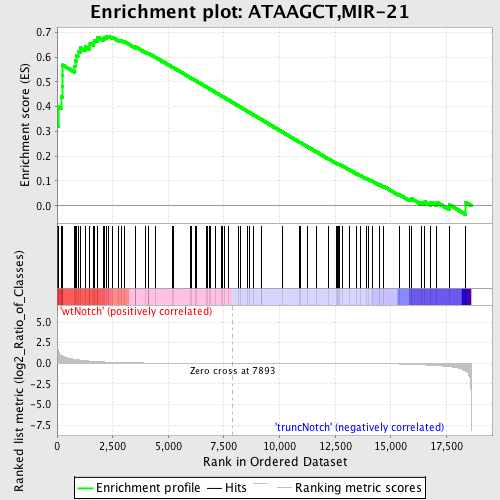
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | ATAAGCT,MIR-21 |
| Enrichment Score (ES) | 0.6843643 |
| Normalized Enrichment Score (NES) | 1.7099173 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.014752251 |
| FWER p-Value | 0.09 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 2630022 1690551 4810110 | 0 | 6.327 | 0.3225 | Yes | ||
| 2 | HNRPK | 1190348 2060411 2570092 6840551 | 56 | 1.549 | 0.3985 | Yes | ||
| 3 | SATB1 | 5670154 | 178 | 0.951 | 0.4404 | Yes | ||
| 4 | MBNL1 | 2640762 7100048 | 235 | 0.865 | 0.4815 | Yes | ||
| 5 | BRD1 | 1660278 | 241 | 0.856 | 0.5248 | Yes | ||
| 6 | PDCD4 | 3360403 5690110 | 246 | 0.850 | 0.5679 | Yes | ||
| 7 | FUBP1 | 5390373 | 784 | 0.477 | 0.5633 | Yes | ||
| 8 | SFRS3 | 770315 4230593 | 810 | 0.468 | 0.5858 | Yes | ||
| 9 | PELI1 | 3870215 6900040 | 881 | 0.439 | 0.6044 | Yes | ||
| 10 | RNF103 | 840452 | 940 | 0.419 | 0.6226 | Yes | ||
| 11 | UBE2D3 | 3190452 | 1032 | 0.381 | 0.6371 | Yes | ||
| 12 | OSR1 | 1500025 | 1264 | 0.319 | 0.6409 | Yes | ||
| 13 | RASGRP1 | 1050148 4050632 6510671 | 1473 | 0.261 | 0.6430 | Yes | ||
| 14 | MTAP | 2120136 | 1476 | 0.261 | 0.6562 | Yes | ||
| 15 | STAT3 | 460040 3710341 | 1643 | 0.227 | 0.6588 | Yes | ||
| 16 | RAB11A | 5360079 | 1684 | 0.220 | 0.6678 | Yes | ||
| 17 | EIF2C2 | 770451 | 1811 | 0.197 | 0.6711 | Yes | ||
| 18 | TGFBR2 | 1780711 1980537 6550398 | 1826 | 0.195 | 0.6802 | Yes | ||
| 19 | RPS6KA3 | 1980707 | 2067 | 0.158 | 0.6753 | Yes | ||
| 20 | KRIT1 | 2640500 4230435 4730184 | 2131 | 0.149 | 0.6795 | Yes | ||
| 21 | PURB | 5360138 | 2214 | 0.136 | 0.6820 | Yes | ||
| 22 | PCBP2 | 5690048 | 2292 | 0.127 | 0.6844 | Yes | ||
| 23 | CDC25A | 3800184 | 2485 | 0.107 | 0.6795 | No | ||
| 24 | STAG2 | 4540132 | 2779 | 0.084 | 0.6679 | No | ||
| 25 | TIMP3 | 1450504 1980270 | 2872 | 0.077 | 0.6669 | No | ||
| 26 | RNF111 | 2970072 3140112 5130647 | 3006 | 0.067 | 0.6631 | No | ||
| 27 | PPP3CA | 4760332 6760092 | 3511 | 0.042 | 0.6380 | No | ||
| 28 | SFRS8 | 5130162 5700164 | 3532 | 0.040 | 0.6390 | No | ||
| 29 | RECK | 840079 | 3535 | 0.040 | 0.6410 | No | ||
| 30 | CART1 | 770338 | 3974 | 0.027 | 0.6187 | No | ||
| 31 | PCBP1 | 1050088 | 4088 | 0.025 | 0.6139 | No | ||
| 32 | ACBD5 | 3440397 6020600 | 4090 | 0.024 | 0.6151 | No | ||
| 33 | SSFA2 | 3870133 | 4128 | 0.024 | 0.6143 | No | ||
| 34 | MRPL9 | 580079 | 4427 | 0.019 | 0.5992 | No | ||
| 35 | GPR64 | 4610692 5220471 | 5169 | 0.011 | 0.5598 | No | ||
| 36 | SCML2 | 3290603 | 5238 | 0.011 | 0.5566 | No | ||
| 37 | CBX4 | 70332 | 5993 | 0.006 | 0.5163 | No | ||
| 38 | NF2 | 4150735 6450139 | 6046 | 0.006 | 0.5138 | No | ||
| 39 | EPHA4 | 460750 | 6208 | 0.005 | 0.5054 | No | ||
| 40 | THRB | 1780673 | 6253 | 0.005 | 0.5033 | No | ||
| 41 | TRPM7 | 2510408 | 6704 | 0.004 | 0.4792 | No | ||
| 42 | SPRY2 | 5860184 | 6779 | 0.003 | 0.4754 | No | ||
| 43 | FASLG | 2810044 | 6871 | 0.003 | 0.4706 | No | ||
| 44 | PLAG1 | 1450142 3870139 6520039 | 6908 | 0.003 | 0.4688 | No | ||
| 45 | CASKIN1 | 580035 3710500 | 7117 | 0.002 | 0.4577 | No | ||
| 46 | PITX2 | 870537 2690139 | 7394 | 0.001 | 0.4429 | No | ||
| 47 | SMAD7 | 430377 | 7414 | 0.001 | 0.4419 | No | ||
| 48 | GLCCI1 | 110091 2640082 2900692 3290070 6370014 | 7530 | 0.001 | 0.4358 | No | ||
| 49 | DAZL | 4050082 | 7683 | 0.001 | 0.4276 | No | ||
| 50 | PPARA | 2060026 | 8143 | -0.001 | 0.4029 | No | ||
| 51 | ASPN | 2450021 | 8248 | -0.001 | 0.3973 | No | ||
| 52 | UBE4A | 6100520 | 8540 | -0.002 | 0.3817 | No | ||
| 53 | BTBD3 | 1980041 2510577 3190053 4200746 | 8628 | -0.002 | 0.3771 | No | ||
| 54 | PPP1R3A | 6760288 | 8812 | -0.003 | 0.3674 | No | ||
| 55 | PIK3R1 | 4730671 | 9200 | -0.004 | 0.3467 | No | ||
| 56 | ZCCHC3 | 580438 1990035 4780278 | 9201 | -0.004 | 0.3469 | No | ||
| 57 | JAG1 | 3440390 | 10130 | -0.007 | 0.2971 | No | ||
| 58 | BNC2 | 4810603 | 10884 | -0.010 | 0.2570 | No | ||
| 59 | CPEB3 | 3940164 | 10953 | -0.010 | 0.2538 | No | ||
| 60 | SOX6 | 2940458 4070601 6840717 | 11235 | -0.011 | 0.2392 | No | ||
| 61 | NFIB | 460450 | 11662 | -0.013 | 0.2169 | No | ||
| 62 | ELF2 | 1690014 2480132 3060180 3190504 | 12215 | -0.017 | 0.1880 | No | ||
| 63 | YAP1 | 4230577 | 12539 | -0.020 | 0.1716 | No | ||
| 64 | MATN2 | 460215 | 12597 | -0.021 | 0.1696 | No | ||
| 65 | CNTFR | 4480092 6200064 | 12654 | -0.021 | 0.1676 | No | ||
| 66 | CCL1 | 4230167 | 12684 | -0.021 | 0.1672 | No | ||
| 67 | NTF3 | 5890131 | 12808 | -0.023 | 0.1617 | No | ||
| 68 | SOX2 | 630487 2230491 | 13164 | -0.027 | 0.1439 | No | ||
| 69 | PER2 | 3120204 3390593 | 13477 | -0.032 | 0.1287 | No | ||
| 70 | ING3 | 110438 2060390 | 13616 | -0.034 | 0.1230 | No | ||
| 71 | JPH1 | 610739 | 13903 | -0.039 | 0.1095 | No | ||
| 72 | KCNA3 | 510059 | 13995 | -0.041 | 0.1067 | No | ||
| 73 | SOX5 | 2370576 2900167 3190128 5050528 | 14189 | -0.045 | 0.0986 | No | ||
| 74 | PCSK6 | 520097 | 14508 | -0.054 | 0.0842 | No | ||
| 75 | SPRY1 | 450253 460093 | 14686 | -0.059 | 0.0777 | No | ||
| 76 | BOLL | 6100743 | 15389 | -0.092 | 0.0445 | No | ||
| 77 | CRIM1 | 130491 | 15853 | -0.126 | 0.0259 | No | ||
| 78 | SKI | 6590008 | 15919 | -0.132 | 0.0291 | No | ||
| 79 | EHD1 | 3840731 | 16384 | -0.186 | 0.0136 | No | ||
| 80 | ARHGAP24 | 540278 4540025 | 16533 | -0.205 | 0.0160 | No | ||
| 81 | TAGAP | 3290433 | 16791 | -0.242 | 0.0145 | No | ||
| 82 | RHOB | 1500309 | 17054 | -0.283 | 0.0148 | No | ||
| 83 | CHD7 | 3870372 | 17618 | -0.403 | 0.0050 | No | ||
| 84 | TGFBI | 2060446 6900112 | 18366 | -0.957 | 0.0135 | No |