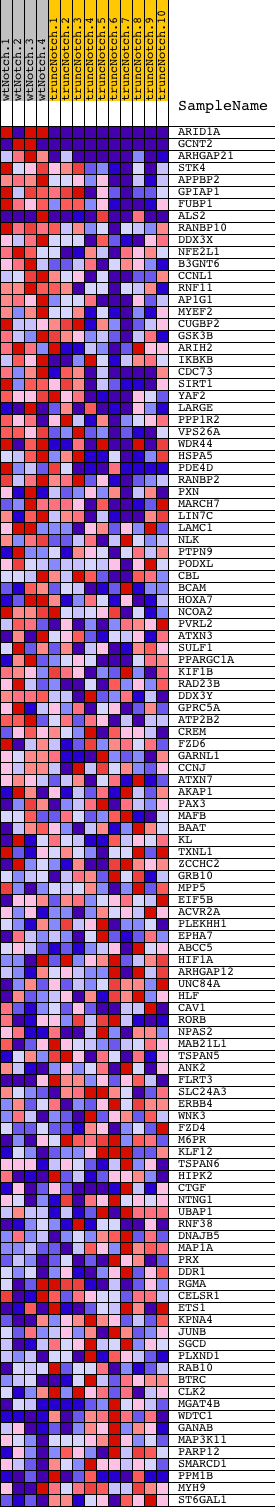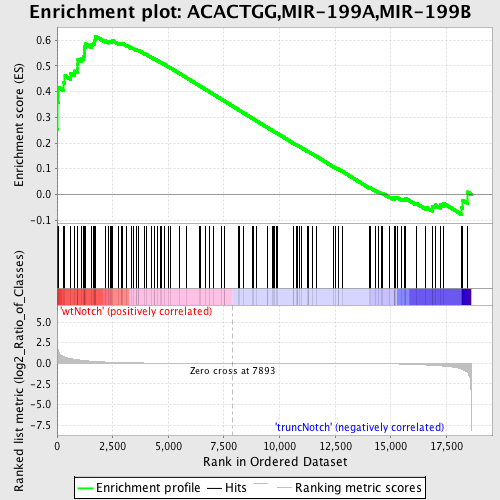
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | ACACTGG,MIR-199A,MIR-199B |
| Enrichment Score (ES) | 0.61607265 |
| Normalized Enrichment Score (NES) | 1.627427 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.032991596 |
| FWER p-Value | 0.322 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 2630022 1690551 4810110 | 0 | 6.327 | 0.2541 | Yes | ||
| 2 | GCNT2 | 6400162 2370609 2760309 4540465 6100280 | 8 | 2.668 | 0.3609 | Yes | ||
| 3 | ARHGAP21 | 2100168 | 64 | 1.460 | 0.4166 | Yes | ||
| 4 | STK4 | 2640152 | 302 | 0.781 | 0.4352 | Yes | ||
| 5 | APPBP2 | 5130215 | 353 | 0.732 | 0.4619 | Yes | ||
| 6 | GPIAP1 | 780551 6420079 | 608 | 0.561 | 0.4706 | Yes | ||
| 7 | FUBP1 | 5390373 | 784 | 0.477 | 0.4804 | Yes | ||
| 8 | ALS2 | 3130546 6400070 | 900 | 0.431 | 0.4915 | Yes | ||
| 9 | RANBP10 | 2340184 | 920 | 0.424 | 0.5075 | Yes | ||
| 10 | DDX3X | 2190020 | 934 | 0.421 | 0.5237 | Yes | ||
| 11 | NFE2L1 | 6130053 | 1084 | 0.365 | 0.5303 | Yes | ||
| 12 | B3GNT6 | 1340736 | 1185 | 0.339 | 0.5385 | Yes | ||
| 13 | CCNL1 | 130273 | 1215 | 0.331 | 0.5502 | Yes | ||
| 14 | RNF11 | 3990068 5080458 | 1231 | 0.327 | 0.5626 | Yes | ||
| 15 | AP1G1 | 2360671 | 1246 | 0.324 | 0.5748 | Yes | ||
| 16 | MYEF2 | 6860484 | 1278 | 0.316 | 0.5858 | Yes | ||
| 17 | CUGBP2 | 6180121 6840139 | 1530 | 0.249 | 0.5823 | Yes | ||
| 18 | GSK3B | 5360348 | 1613 | 0.232 | 0.5871 | Yes | ||
| 19 | ARIH2 | 2810025 | 1673 | 0.222 | 0.5929 | Yes | ||
| 20 | IKBKB | 6840072 | 1694 | 0.218 | 0.6005 | Yes | ||
| 21 | CDC73 | 1580132 | 1708 | 0.216 | 0.6085 | Yes | ||
| 22 | SIRT1 | 1190731 | 1727 | 0.212 | 0.6161 | Yes | ||
| 23 | YAF2 | 940053 | 2175 | 0.141 | 0.5976 | No | ||
| 24 | LARGE | 6940068 | 2331 | 0.123 | 0.5942 | No | ||
| 25 | PPP1R2 | 3710278 1400725 | 2402 | 0.116 | 0.5950 | No | ||
| 26 | VPS26A | 1940494 5720059 | 2449 | 0.111 | 0.5970 | No | ||
| 27 | WDR44 | 130195 1050609 1570685 | 2479 | 0.108 | 0.5998 | No | ||
| 28 | HSPA5 | 5130671 | 2757 | 0.085 | 0.5882 | No | ||
| 29 | PDE4D | 2470528 6660014 | 2880 | 0.076 | 0.5847 | No | ||
| 30 | RANBP2 | 4280338 | 2886 | 0.075 | 0.5874 | No | ||
| 31 | PXN | 3290048 6400132 | 2950 | 0.071 | 0.5869 | No | ||
| 32 | MARCH7 | 3800020 6620168 | 3128 | 0.060 | 0.5797 | No | ||
| 33 | LIN7C | 4850341 | 3344 | 0.048 | 0.5700 | No | ||
| 34 | LAMC1 | 4730025 | 3413 | 0.045 | 0.5682 | No | ||
| 35 | NLK | 2030010 2450041 | 3545 | 0.040 | 0.5627 | No | ||
| 36 | PTPN9 | 3290408 | 3585 | 0.039 | 0.5621 | No | ||
| 37 | PODXL | 130717 | 3635 | 0.037 | 0.5610 | No | ||
| 38 | CBL | 6380068 | 3650 | 0.036 | 0.5616 | No | ||
| 39 | BCAM | 70022 | 3947 | 0.027 | 0.5467 | No | ||
| 40 | HOXA7 | 5910152 | 4020 | 0.026 | 0.5439 | No | ||
| 41 | NCOA2 | 940347 2850725 | 4223 | 0.022 | 0.5339 | No | ||
| 42 | PVRL2 | 2760131 4050114 | 4356 | 0.020 | 0.5275 | No | ||
| 43 | ATXN3 | 50156 2900095 | 4389 | 0.020 | 0.5266 | No | ||
| 44 | SULF1 | 430575 | 4529 | 0.018 | 0.5198 | No | ||
| 45 | PPARGC1A | 4670040 | 4653 | 0.016 | 0.5138 | No | ||
| 46 | KIF1B | 1240494 2370139 4570270 6510102 | 4683 | 0.016 | 0.5129 | No | ||
| 47 | RAD23B | 2190671 | 4843 | 0.014 | 0.5048 | No | ||
| 48 | DDX3Y | 1580278 4200519 | 4985 | 0.013 | 0.4977 | No | ||
| 49 | GPRC5A | 3870095 | 5080 | 0.012 | 0.4931 | No | ||
| 50 | ATP2B2 | 3780397 | 5480 | 0.009 | 0.4719 | No | ||
| 51 | CREM | 840156 6380438 6660041 6660168 | 5828 | 0.007 | 0.4535 | No | ||
| 52 | FZD6 | 360288 | 6409 | 0.005 | 0.4223 | No | ||
| 53 | GARNL1 | 5670398 | 6459 | 0.004 | 0.4198 | No | ||
| 54 | CCNJ | 870047 | 6682 | 0.004 | 0.4080 | No | ||
| 55 | ATXN7 | 1450435 | 6868 | 0.003 | 0.3981 | No | ||
| 56 | AKAP1 | 110148 1740735 2260019 7000563 | 7026 | 0.003 | 0.3897 | No | ||
| 57 | PAX3 | 50551 | 7370 | 0.001 | 0.3712 | No | ||
| 58 | MAFB | 1230471 | 7513 | 0.001 | 0.3636 | No | ||
| 59 | BAAT | 730739 | 7518 | 0.001 | 0.3634 | No | ||
| 60 | KL | 3120471 | 8155 | -0.001 | 0.3291 | No | ||
| 61 | TXNL1 | 2850148 | 8191 | -0.001 | 0.3272 | No | ||
| 62 | ZCCHC2 | 460592 | 8364 | -0.001 | 0.3180 | No | ||
| 63 | GRB10 | 6980082 | 8776 | -0.002 | 0.2959 | No | ||
| 64 | MPP5 | 2100148 5220632 | 8826 | -0.003 | 0.2933 | No | ||
| 65 | EIF5B | 1190102 4200440 | 8972 | -0.003 | 0.2856 | No | ||
| 66 | ACVR2A | 6110647 | 9461 | -0.004 | 0.2594 | No | ||
| 67 | PLEKHH1 | 4780280 | 9462 | -0.004 | 0.2596 | No | ||
| 68 | EPHA7 | 1190288 4010278 | 9702 | -0.005 | 0.2469 | No | ||
| 69 | ABCC5 | 2100600 5050692 | 9721 | -0.005 | 0.2461 | No | ||
| 70 | HIF1A | 5670605 | 9774 | -0.005 | 0.2435 | No | ||
| 71 | ARHGAP12 | 1050181 | 9846 | -0.006 | 0.2399 | No | ||
| 72 | UNC84A | 1450632 3520408 | 9904 | -0.006 | 0.2371 | No | ||
| 73 | HLF | 2370113 | 10610 | -0.008 | 0.1993 | No | ||
| 74 | CAV1 | 870025 | 10616 | -0.008 | 0.1994 | No | ||
| 75 | RORB | 6400035 | 10759 | -0.009 | 0.1921 | No | ||
| 76 | NPAS2 | 770670 | 10812 | -0.009 | 0.1896 | No | ||
| 77 | MAB21L1 | 1450634 1580092 | 10816 | -0.009 | 0.1898 | No | ||
| 78 | TSPAN5 | 6770019 | 10898 | -0.010 | 0.1858 | No | ||
| 79 | ANK2 | 6510546 | 10963 | -0.010 | 0.1828 | No | ||
| 80 | FLRT3 | 70685 2760497 6040519 | 11275 | -0.011 | 0.1664 | No | ||
| 81 | SLC24A3 | 5080707 | 11279 | -0.011 | 0.1667 | No | ||
| 82 | ERBB4 | 4610400 4810017 | 11308 | -0.012 | 0.1657 | No | ||
| 83 | WNK3 | 4060053 | 11477 | -0.012 | 0.1571 | No | ||
| 84 | FZD4 | 1580520 | 11677 | -0.014 | 0.1469 | No | ||
| 85 | M6PR | 2480458 3290593 3840324 | 12436 | -0.019 | 0.1067 | No | ||
| 86 | KLF12 | 1660095 4810288 5340546 6520286 | 12491 | -0.020 | 0.1046 | No | ||
| 87 | TSPAN6 | 2470201 3520358 | 12626 | -0.021 | 0.0981 | No | ||
| 88 | HIPK2 | 6350647 | 12627 | -0.021 | 0.0990 | No | ||
| 89 | CTGF | 4540577 | 12821 | -0.023 | 0.0895 | No | ||
| 90 | NTNG1 | 50270 1410427 4810333 | 14060 | -0.042 | 0.0243 | No | ||
| 91 | UBAP1 | 4280059 | 14067 | -0.043 | 0.0257 | No | ||
| 92 | RNF38 | 1940746 3140133 3840601 | 14311 | -0.048 | 0.0144 | No | ||
| 93 | DNAJB5 | 2900215 | 14457 | -0.052 | 0.0087 | No | ||
| 94 | MAP1A | 4920576 | 14569 | -0.056 | 0.0050 | No | ||
| 95 | PRX | 2260400 4200347 | 14606 | -0.057 | 0.0053 | No | ||
| 96 | DDR1 | 2060044 5220180 | 14951 | -0.069 | -0.0105 | No | ||
| 97 | RGMA | 2450711 | 15151 | -0.079 | -0.0181 | No | ||
| 98 | CELSR1 | 6510348 | 15158 | -0.079 | -0.0153 | No | ||
| 99 | ETS1 | 5270278 6450717 6620465 | 15170 | -0.079 | -0.0127 | No | ||
| 100 | KPNA4 | 510369 | 15210 | -0.081 | -0.0115 | No | ||
| 101 | JUNB | 4230048 | 15318 | -0.088 | -0.0137 | No | ||
| 102 | SGCD | 2940685 5080753 | 15477 | -0.098 | -0.0183 | No | ||
| 103 | PLXND1 | 5270682 | 15610 | -0.106 | -0.0212 | No | ||
| 104 | RAB10 | 2360008 5890020 | 15646 | -0.108 | -0.0187 | No | ||
| 105 | BTRC | 3170131 | 15653 | -0.109 | -0.0147 | No | ||
| 106 | CLK2 | 510079 2630736 | 16145 | -0.156 | -0.0350 | No | ||
| 107 | MGAT4B | 130053 6840100 | 16570 | -0.210 | -0.0495 | No | ||
| 108 | WDTC1 | 1690541 | 16856 | -0.253 | -0.0547 | No | ||
| 109 | GANAB | 1980400 5720717 | 16875 | -0.255 | -0.0455 | No | ||
| 110 | MAP3K11 | 7000039 | 16996 | -0.273 | -0.0410 | No | ||
| 111 | PARP12 | 4810152 | 17224 | -0.317 | -0.0405 | No | ||
| 112 | SMARCD1 | 3060193 3850184 6400369 | 17376 | -0.347 | -0.0348 | No | ||
| 113 | PPM1B | 380180 450154 3610215 | 18181 | -0.684 | -0.0508 | No | ||
| 114 | MYH9 | 3120091 4850292 | 18234 | -0.754 | -0.0233 | No | ||
| 115 | ST6GAL1 | 2970546 | 18443 | -1.093 | 0.0094 | No |