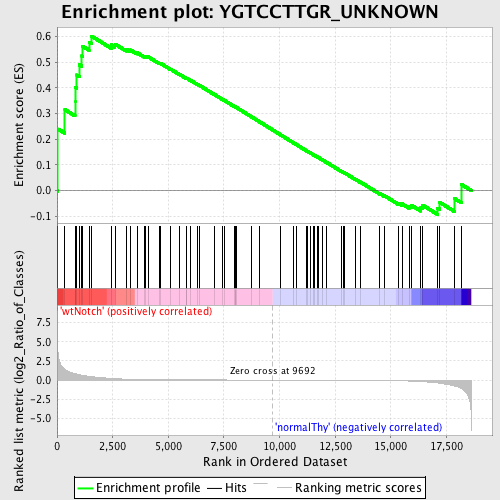
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | YGTCCTTGR_UNKNOWN |
| Enrichment Score (ES) | 0.60064626 |
| Normalized Enrichment Score (NES) | 1.3129773 |
| Nominal p-value | 0.08188153 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPP2R4 | 670280 2900097 | 37 | 3.665 | 0.2396 | Yes | ||
| 2 | ALDOA | 6290672 | 351 | 1.397 | 0.3148 | Yes | ||
| 3 | CYCS | 2030072 | 807 | 0.851 | 0.3464 | Yes | ||
| 4 | ATP2A2 | 1090075 3990279 | 812 | 0.849 | 0.4021 | Yes | ||
| 5 | PRKACA | 2640731 4050048 | 889 | 0.782 | 0.4496 | Yes | ||
| 6 | OTX2 | 1190471 | 999 | 0.685 | 0.4889 | Yes | ||
| 7 | HIST1H1A | 1450131 | 1113 | 0.618 | 0.5235 | Yes | ||
| 8 | ESM1 | 1410594 | 1143 | 0.604 | 0.5617 | Yes | ||
| 9 | SYT13 | 5910110 | 1438 | 0.484 | 0.5778 | Yes | ||
| 10 | KCNAB3 | 2470725 | 1550 | 0.438 | 0.6006 | Yes | ||
| 11 | CS | 5080600 | 2436 | 0.210 | 0.5668 | No | ||
| 12 | GAS6 | 4480021 | 2602 | 0.179 | 0.5697 | No | ||
| 13 | PRKAG1 | 6450091 | 3107 | 0.120 | 0.5505 | No | ||
| 14 | TMEM16D | 2650324 | 3300 | 0.101 | 0.5468 | No | ||
| 15 | CTRL | 2120301 | 3594 | 0.078 | 0.5362 | No | ||
| 16 | SNTA1 | 4570400 | 3948 | 0.057 | 0.5209 | No | ||
| 17 | THRA | 6770341 110164 1990600 | 3973 | 0.056 | 0.5233 | No | ||
| 18 | PANK3 | 3060687 | 4092 | 0.050 | 0.5202 | No | ||
| 19 | ARMET | 5130670 | 4591 | 0.036 | 0.4957 | No | ||
| 20 | SOCS5 | 3830398 7100093 | 4649 | 0.034 | 0.4949 | No | ||
| 21 | ADAM11 | 1050008 3130494 | 5104 | 0.026 | 0.4721 | No | ||
| 22 | CLCNKA | 1740050 | 5519 | 0.021 | 0.4512 | No | ||
| 23 | YWHAG | 3780341 | 5797 | 0.018 | 0.4374 | No | ||
| 24 | RGL1 | 1500097 6900450 | 5824 | 0.018 | 0.4372 | No | ||
| 25 | AMY2A | 580138 | 5983 | 0.016 | 0.4297 | No | ||
| 26 | HMGCS1 | 6620452 | 6288 | 0.014 | 0.4143 | No | ||
| 27 | MLLT6 | 3870168 | 6398 | 0.013 | 0.4093 | No | ||
| 28 | NPTX2 | 70021 | 7090 | 0.009 | 0.3726 | No | ||
| 29 | TNKS1BP1 | 7040184 | 7447 | 0.008 | 0.3540 | No | ||
| 30 | GABRA3 | 4810142 | 7541 | 0.007 | 0.3494 | No | ||
| 31 | MT3 | 1450537 | 7976 | 0.006 | 0.3264 | No | ||
| 32 | CBX4 | 70332 | 8001 | 0.006 | 0.3255 | No | ||
| 33 | HOXD11 | 2260242 | 8020 | 0.006 | 0.3249 | No | ||
| 34 | EMX2 | 1660092 | 8041 | 0.005 | 0.3242 | No | ||
| 35 | SNCB | 3360133 | 8733 | 0.003 | 0.2871 | No | ||
| 36 | CITED2 | 5670114 5130088 | 9116 | 0.002 | 0.2667 | No | ||
| 37 | TRIM2 | 430546 1170286 | 10027 | -0.001 | 0.2177 | No | ||
| 38 | IGSF3 | 6450524 | 10612 | -0.003 | 0.1864 | No | ||
| 39 | ZIC3 | 380020 | 10740 | -0.003 | 0.1797 | No | ||
| 40 | PRKCE | 5700053 | 11223 | -0.005 | 0.1541 | No | ||
| 41 | SYNPO2L | 5390719 | 11243 | -0.005 | 0.1534 | No | ||
| 42 | SYT7 | 2450528 3390500 3610687 | 11368 | -0.005 | 0.1470 | No | ||
| 43 | PAM | 5290528 | 11508 | -0.006 | 0.1399 | No | ||
| 44 | DRD3 | 4780402 | 11570 | -0.006 | 0.1370 | No | ||
| 45 | CHST8 | 1940050 | 11713 | -0.007 | 0.1298 | No | ||
| 46 | KCNJ16 | 4780273 | 11750 | -0.007 | 0.1283 | No | ||
| 47 | ELAVL2 | 360181 | 11935 | -0.008 | 0.1189 | No | ||
| 48 | CRAT | 540020 2060364 3520148 | 12105 | -0.008 | 0.1104 | No | ||
| 49 | SEMA6D | 4050324 5860138 6350307 | 12768 | -0.012 | 0.0755 | No | ||
| 50 | ARPC5 | 1740411 3450300 | 12894 | -0.013 | 0.0696 | No | ||
| 51 | AFP | 6590358 | 12932 | -0.013 | 0.0684 | No | ||
| 52 | MCAM | 5130270 6620577 | 13416 | -0.017 | 0.0436 | No | ||
| 53 | PHYHIPL | 2360706 3840692 | 13626 | -0.020 | 0.0336 | No | ||
| 54 | HRH3 | 6180040 | 14504 | -0.035 | -0.0114 | No | ||
| 55 | SLC38A2 | 1470242 3800026 | 14697 | -0.041 | -0.0190 | No | ||
| 56 | H2AFZ | 1470168 | 15338 | -0.075 | -0.0485 | No | ||
| 57 | VAMP2 | 2100100 | 15502 | -0.091 | -0.0513 | No | ||
| 58 | FHL2 | 1400008 | 15820 | -0.124 | -0.0602 | No | ||
| 59 | ATP5B | 3870138 | 15922 | -0.137 | -0.0566 | No | ||
| 60 | DIRC2 | 2810494 | 16348 | -0.201 | -0.0663 | No | ||
| 61 | HDAC3 | 4060072 | 16425 | -0.215 | -0.0562 | No | ||
| 62 | CENTD1 | 2570592 | 17107 | -0.376 | -0.0681 | No | ||
| 63 | BAZ2A | 730184 | 17202 | -0.414 | -0.0458 | No | ||
| 64 | HSPBAP1 | 4060022 | 17865 | -0.763 | -0.0312 | No | ||
| 65 | STIM1 | 6380138 | 18164 | -1.087 | 0.0244 | No |

