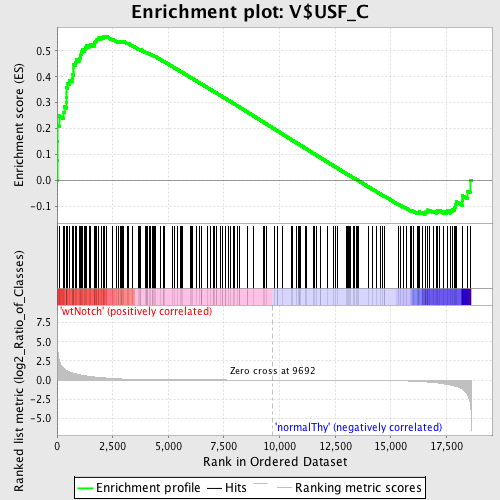
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$USF_C |
| Enrichment Score (ES) | 0.5567144 |
| Normalized Enrichment Score (NES) | 1.365518 |
| Nominal p-value | 0.016447369 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PABPC1 | 2650180 2690253 6020632 1990270 | 6 | 5.043 | 0.0789 | Yes | ||
| 2 | TIMM10 | 610070 1450019 | 20 | 4.600 | 0.1505 | Yes | ||
| 3 | PABPC4 | 1990170 6760270 5390138 | 33 | 3.900 | 0.2111 | Yes | ||
| 4 | HMOX1 | 1740687 | 92 | 2.622 | 0.2492 | Yes | ||
| 5 | ENO3 | 5270136 | 302 | 1.532 | 0.2619 | Yes | ||
| 6 | AKAP12 | 1450739 | 321 | 1.471 | 0.2840 | Yes | ||
| 7 | ABCB6 | 6350138 | 409 | 1.281 | 0.2994 | Yes | ||
| 8 | SH3KBP1 | 670102 2850458 7000309 | 415 | 1.267 | 0.3191 | Yes | ||
| 9 | IPO4 | 5050619 | 419 | 1.251 | 0.3386 | Yes | ||
| 10 | FBL | 5130020 | 420 | 1.249 | 0.3582 | Yes | ||
| 11 | LYAR | 7000402 | 461 | 1.171 | 0.3744 | Yes | ||
| 12 | G6PC3 | 1580487 5390253 | 540 | 1.061 | 0.3869 | Yes | ||
| 13 | RPS19 | 5860066 | 672 | 0.943 | 0.3946 | Yes | ||
| 14 | EXOSC5 | 3370671 | 686 | 0.933 | 0.4085 | Yes | ||
| 15 | PFDN2 | 4230156 | 738 | 0.895 | 0.4198 | Yes | ||
| 16 | PPAT | 1980019 | 740 | 0.895 | 0.4338 | Yes | ||
| 17 | PA2G4 | 2030463 | 742 | 0.894 | 0.4478 | Yes | ||
| 18 | EXOSC2 | 6900471 | 832 | 0.827 | 0.4560 | Yes | ||
| 19 | GTF2H1 | 2570746 2650148 | 855 | 0.811 | 0.4676 | Yes | ||
| 20 | NUDC | 4780025 | 986 | 0.691 | 0.4714 | Yes | ||
| 21 | HYAL2 | 1500152 | 1034 | 0.665 | 0.4793 | Yes | ||
| 22 | SDC1 | 3440471 6350408 | 1070 | 0.645 | 0.4875 | Yes | ||
| 23 | OPRS1 | 6130128 6290717 | 1100 | 0.628 | 0.4958 | Yes | ||
| 24 | FKBP11 | 3170575 | 1126 | 0.610 | 0.5040 | Yes | ||
| 25 | HMGN2 | 3140091 | 1250 | 0.558 | 0.5061 | Yes | ||
| 26 | EIF4B | 5390563 5270577 | 1274 | 0.548 | 0.5135 | Yes | ||
| 27 | EIF2C2 | 770451 | 1325 | 0.528 | 0.5191 | Yes | ||
| 28 | GPD1 | 2480095 | 1435 | 0.484 | 0.5208 | Yes | ||
| 29 | PAICS | 610142 | 1511 | 0.452 | 0.5238 | Yes | ||
| 30 | HSPE1 | 3170438 | 1661 | 0.403 | 0.5220 | Yes | ||
| 31 | RPL22 | 4070070 | 1675 | 0.397 | 0.5276 | Yes | ||
| 32 | IVNS1ABP | 4760601 6520113 | 1688 | 0.393 | 0.5331 | Yes | ||
| 33 | MORF4L2 | 6450133 | 1725 | 0.384 | 0.5372 | Yes | ||
| 34 | PSMB3 | 4280594 | 1768 | 0.372 | 0.5407 | Yes | ||
| 35 | SRP72 | 3990427 | 1789 | 0.367 | 0.5454 | Yes | ||
| 36 | POLR3E | 380152 | 1855 | 0.347 | 0.5473 | Yes | ||
| 37 | HERPUD1 | 7100440 | 1881 | 0.340 | 0.5513 | Yes | ||
| 38 | ANXA6 | 2190014 | 1991 | 0.311 | 0.5503 | Yes | ||
| 39 | EIF4G1 | 4070446 | 2065 | 0.292 | 0.5509 | Yes | ||
| 40 | FHOD1 | 2680279 | 2097 | 0.283 | 0.5537 | Yes | ||
| 41 | BCL9L | 6940053 | 2123 | 0.279 | 0.5567 | Yes | ||
| 42 | MYOHD1 | 1410520 | 2220 | 0.256 | 0.5555 | No | ||
| 43 | POLR2H | 5890242 | 2477 | 0.201 | 0.5448 | No | ||
| 44 | SLC39A7 | 5570131 | 2659 | 0.171 | 0.5377 | No | ||
| 45 | MPP3 | 1400056 4480411 6770168 | 2743 | 0.159 | 0.5357 | No | ||
| 46 | MYST3 | 5270500 | 2753 | 0.158 | 0.5377 | No | ||
| 47 | STMN1 | 1990717 | 2854 | 0.145 | 0.5345 | No | ||
| 48 | CACNA1D | 1740315 4730731 | 2891 | 0.140 | 0.5348 | No | ||
| 49 | PSEN2 | 130382 | 2901 | 0.139 | 0.5364 | No | ||
| 50 | PPARGC1B | 3290114 | 2930 | 0.136 | 0.5371 | No | ||
| 51 | CDC14A | 4050132 | 2962 | 0.133 | 0.5375 | No | ||
| 52 | B4GALT2 | 780037 | 3150 | 0.115 | 0.5291 | No | ||
| 53 | UBE1 | 4150139 7050463 | 3187 | 0.112 | 0.5290 | No | ||
| 54 | TIMM9 | 2450484 | 3376 | 0.094 | 0.5202 | No | ||
| 55 | CNNM1 | 2190433 | 3649 | 0.074 | 0.5066 | No | ||
| 56 | RUNX2 | 1400193 4230215 | 3699 | 0.070 | 0.5051 | No | ||
| 57 | FARP1 | 5290102 7100341 | 3744 | 0.067 | 0.5038 | No | ||
| 58 | AMPD2 | 630093 1170070 | 3769 | 0.066 | 0.5035 | No | ||
| 59 | HSPH1 | 1690017 | 3981 | 0.055 | 0.4929 | No | ||
| 60 | SLC26A2 | 2360278 | 4005 | 0.054 | 0.4925 | No | ||
| 61 | SYT3 | 6380017 | 4014 | 0.053 | 0.4929 | No | ||
| 62 | TNFRSF21 | 6380100 | 4046 | 0.052 | 0.4921 | No | ||
| 63 | ARRDC3 | 4610390 | 4077 | 0.051 | 0.4912 | No | ||
| 64 | BHLHB2 | 7040603 | 4158 | 0.048 | 0.4876 | No | ||
| 65 | TIAL1 | 4150048 6510605 | 4191 | 0.046 | 0.4866 | No | ||
| 66 | NLN | 1450576 5870575 | 4277 | 0.044 | 0.4827 | No | ||
| 67 | UBXD3 | 5050193 | 4291 | 0.043 | 0.4827 | No | ||
| 68 | SUCLG2 | 5340397 5700270 | 4316 | 0.042 | 0.4820 | No | ||
| 69 | CAMK4 | 1690091 | 4383 | 0.041 | 0.4791 | No | ||
| 70 | EFNB1 | 2970433 | 4415 | 0.040 | 0.4780 | No | ||
| 71 | SOCS5 | 3830398 7100093 | 4649 | 0.034 | 0.4659 | No | ||
| 72 | ALDH3B1 | 4210010 6940403 | 4764 | 0.032 | 0.4603 | No | ||
| 73 | AVP | 2100113 | 4766 | 0.032 | 0.4607 | No | ||
| 74 | DAZAP1 | 5890563 | 4812 | 0.031 | 0.4587 | No | ||
| 75 | NR1D1 | 2360471 770746 6590204 | 5178 | 0.025 | 0.4393 | No | ||
| 76 | GTF2A1 | 4120138 4920524 5390110 1230397 | 5284 | 0.024 | 0.4340 | No | ||
| 77 | HOXA3 | 3610632 6020358 | 5425 | 0.022 | 0.4267 | No | ||
| 78 | GPRC5C | 1580372 | 5556 | 0.020 | 0.4200 | No | ||
| 79 | SYT6 | 2510280 3850128 4540064 | 5610 | 0.020 | 0.4175 | No | ||
| 80 | SEMA7A | 2630239 5050593 | 5620 | 0.020 | 0.4173 | No | ||
| 81 | HS3ST3A1 | 2230497 | 6008 | 0.016 | 0.3965 | No | ||
| 82 | XRN2 | 130603 1230113 4230309 | 6018 | 0.016 | 0.3963 | No | ||
| 83 | RUSC1 | 6450093 6980575 | 6079 | 0.016 | 0.3933 | No | ||
| 84 | COL2A1 | 460446 3940500 | 6245 | 0.014 | 0.3846 | No | ||
| 85 | PIGW | 4810072 | 6396 | 0.013 | 0.3766 | No | ||
| 86 | SC5DL | 6650390 | 6468 | 0.013 | 0.3730 | No | ||
| 87 | TBX4 | 3060008 | 6471 | 0.013 | 0.3731 | No | ||
| 88 | LTBR | 2450341 | 6494 | 0.013 | 0.3721 | No | ||
| 89 | NXPH1 | 3870546 | 6751 | 0.011 | 0.3584 | No | ||
| 90 | SGTB | 460170 | 6903 | 0.010 | 0.3503 | No | ||
| 91 | FGF14 | 2630390 3520075 6770048 | 7022 | 0.010 | 0.3441 | No | ||
| 92 | ELOVL4 | 5220333 | 7027 | 0.010 | 0.3440 | No | ||
| 93 | KIAA0427 | 1690435 | 7079 | 0.009 | 0.3414 | No | ||
| 94 | DNAJB9 | 460114 | 7148 | 0.009 | 0.3378 | No | ||
| 95 | BMP4 | 380113 | 7150 | 0.009 | 0.3379 | No | ||
| 96 | ETV1 | 70735 2940603 5080463 | 7354 | 0.008 | 0.3270 | No | ||
| 97 | PLA2G4A | 6380364 | 7420 | 0.008 | 0.3236 | No | ||
| 98 | DRD1 | 430025 | 7424 | 0.008 | 0.3236 | No | ||
| 99 | RHEBL1 | 2810576 | 7552 | 0.007 | 0.3168 | No | ||
| 100 | LIN28 | 6590672 | 7554 | 0.007 | 0.3169 | No | ||
| 101 | SLC12A5 | 1980692 | 7708 | 0.007 | 0.3087 | No | ||
| 102 | DNMT3A | 4050068 4810717 | 7784 | 0.006 | 0.3047 | No | ||
| 103 | POGK | 5290315 | 7932 | 0.006 | 0.2968 | No | ||
| 104 | SATB2 | 2470181 | 7962 | 0.006 | 0.2954 | No | ||
| 105 | HOXC12 | 5270411 | 8110 | 0.005 | 0.2875 | No | ||
| 106 | HOXC13 | 4070722 | 8182 | 0.005 | 0.2837 | No | ||
| 107 | BMP2K | 60541 5570504 | 8205 | 0.005 | 0.2826 | No | ||
| 108 | GRIN2A | 6550538 | 8554 | 0.004 | 0.2637 | No | ||
| 109 | CGN | 5890139 6370167 6400537 | 8556 | 0.004 | 0.2637 | No | ||
| 110 | FGF11 | 840292 | 8843 | 0.003 | 0.2483 | No | ||
| 111 | CHRM1 | 4280619 | 9256 | 0.001 | 0.2259 | No | ||
| 112 | TLL1 | 2680315 | 9310 | 0.001 | 0.2231 | No | ||
| 113 | SNX5 | 1570021 6860609 | 9427 | 0.001 | 0.2168 | No | ||
| 114 | LAP3 | 2570064 | 9786 | -0.000 | 0.1974 | No | ||
| 115 | CTSF | 6100021 | 9893 | -0.001 | 0.1916 | No | ||
| 116 | LRP8 | 3610746 5360035 | 9894 | -0.001 | 0.1916 | No | ||
| 117 | SIRT1 | 1190731 | 10151 | -0.001 | 0.1778 | No | ||
| 118 | ARX | 6900504 | 10554 | -0.003 | 0.1560 | No | ||
| 119 | MTHFD1 | 5900398 6520427 | 10571 | -0.003 | 0.1552 | No | ||
| 120 | NKX2-3 | 1990736 6940088 | 10777 | -0.003 | 0.1441 | No | ||
| 121 | LHX9 | 2350112 | 10869 | -0.004 | 0.1392 | No | ||
| 122 | ABCA1 | 6290156 | 10872 | -0.004 | 0.1392 | No | ||
| 123 | FADS3 | 3120079 | 10916 | -0.004 | 0.1369 | No | ||
| 124 | ENPP6 | 110524 580563 | 10940 | -0.004 | 0.1357 | No | ||
| 125 | MAX | 5220021 | 11143 | -0.005 | 0.1248 | No | ||
| 126 | PRKCE | 5700053 | 11223 | -0.005 | 0.1206 | No | ||
| 127 | PAX6 | 1190025 | 11545 | -0.006 | 0.1033 | No | ||
| 128 | SLC6A15 | 5290176 6040017 | 11555 | -0.006 | 0.1029 | No | ||
| 129 | EIF3S1 | 6130368 6770044 | 11678 | -0.006 | 0.0964 | No | ||
| 130 | HPCA | 6860010 | 11854 | -0.007 | 0.0870 | No | ||
| 131 | NPM1 | 4730427 | 11860 | -0.007 | 0.0868 | No | ||
| 132 | FLT3 | 2640487 | 12159 | -0.009 | 0.0708 | No | ||
| 133 | SET | 6650286 | 12420 | -0.010 | 0.0569 | No | ||
| 134 | TDRD1 | 4230301 6110750 | 12534 | -0.011 | 0.0509 | No | ||
| 135 | SCFD2 | 2480504 | 12596 | -0.011 | 0.0478 | No | ||
| 136 | WEE1 | 3390070 | 13018 | -0.014 | 0.0251 | No | ||
| 137 | EEF1E1 | 4540711 | 13040 | -0.014 | 0.0242 | No | ||
| 138 | IPO13 | 3870088 | 13106 | -0.014 | 0.0209 | No | ||
| 139 | HPCAL4 | 2600484 | 13147 | -0.015 | 0.0190 | No | ||
| 140 | DAZL | 4050082 | 13199 | -0.015 | 0.0164 | No | ||
| 141 | NPTX1 | 3710736 | 13341 | -0.017 | 0.0091 | No | ||
| 142 | HHIP | 6940438 | 13385 | -0.017 | 0.0070 | No | ||
| 143 | ADAMTS17 | 6510739 | 13463 | -0.018 | 0.0031 | No | ||
| 144 | SNCAIP | 4010551 | 13516 | -0.018 | 0.0006 | No | ||
| 145 | CHST11 | 6760546 | 13531 | -0.018 | 0.0001 | No | ||
| 146 | ETV4 | 2450605 | 13983 | -0.025 | -0.0240 | No | ||
| 147 | CNOT4 | 2320242 6760706 | 14178 | -0.028 | -0.0341 | No | ||
| 148 | MCM8 | 6130743 | 14349 | -0.032 | -0.0428 | No | ||
| 149 | CLCN2 | 1940021 | 14536 | -0.036 | -0.0523 | No | ||
| 150 | ADCY3 | 2350347 3440242 | 14619 | -0.039 | -0.0562 | No | ||
| 151 | RPL13A | 2680519 5700142 | 14717 | -0.042 | -0.0608 | No | ||
| 152 | PER1 | 3290315 | 14719 | -0.042 | -0.0602 | No | ||
| 153 | HOXA4 | 940152 | 15336 | -0.075 | -0.0924 | No | ||
| 154 | SLC31A2 | 1450577 | 15445 | -0.085 | -0.0969 | No | ||
| 155 | PTMA | 5570148 | 15567 | -0.096 | -0.1020 | No | ||
| 156 | BCKDHA | 50189 | 15686 | -0.108 | -0.1067 | No | ||
| 157 | CRMP1 | 4810014 | 15890 | -0.132 | -0.1156 | No | ||
| 158 | GRK6 | 1500053 | 15906 | -0.135 | -0.1143 | No | ||
| 159 | MNT | 4920575 | 16026 | -0.150 | -0.1184 | No | ||
| 160 | EIF4E | 1580403 70133 6380215 | 16200 | -0.177 | -0.1250 | No | ||
| 161 | RXRB | 1780040 5340438 | 16248 | -0.183 | -0.1247 | No | ||
| 162 | HSPA4 | 1050170 3450309 | 16253 | -0.185 | -0.1220 | No | ||
| 163 | RPL19 | 5550592 | 16277 | -0.188 | -0.1203 | No | ||
| 164 | MAT2A | 4070026 4730079 6020280 | 16421 | -0.215 | -0.1247 | No | ||
| 165 | GPX1 | 4150093 | 16538 | -0.236 | -0.1273 | No | ||
| 166 | USP2 | 1190292 1240253 4850035 | 16553 | -0.238 | -0.1243 | No | ||
| 167 | PDK2 | 2690017 | 16570 | -0.243 | -0.1213 | No | ||
| 168 | MXD4 | 4810113 | 16630 | -0.252 | -0.1206 | No | ||
| 169 | GNA13 | 4590102 | 16639 | -0.254 | -0.1170 | No | ||
| 170 | CENTB5 | 5690128 | 16650 | -0.255 | -0.1135 | No | ||
| 171 | ARF6 | 3520026 | 16734 | -0.270 | -0.1138 | No | ||
| 172 | H3F3A | 2900086 | 16916 | -0.317 | -0.1186 | No | ||
| 173 | PFN1 | 6130132 | 17047 | -0.354 | -0.1201 | No | ||
| 174 | SLC20A1 | 6420014 | 17083 | -0.367 | -0.1163 | No | ||
| 175 | EIF5A | 1500059 5290022 6370026 | 17168 | -0.397 | -0.1146 | No | ||
| 176 | BCAS3 | 4050647 | 17388 | -0.496 | -0.1187 | No | ||
| 177 | FCHSD2 | 5720092 | 17525 | -0.558 | -0.1173 | No | ||
| 178 | TEF | 1050050 3120397 4060487 | 17678 | -0.631 | -0.1156 | No | ||
| 179 | ACVR2B | 1660546 | 17778 | -0.700 | -0.1100 | No | ||
| 180 | HSPBAP1 | 4060022 | 17865 | -0.763 | -0.1026 | No | ||
| 181 | GABARAP | 1450286 | 17920 | -0.802 | -0.0930 | No | ||
| 182 | DIABLO | 2360170 | 17935 | -0.815 | -0.0809 | No | ||
| 183 | PPP1R9B | 3130619 | 18215 | -1.187 | -0.0774 | No | ||
| 184 | WBP2 | 3170128 | 18224 | -1.201 | -0.0590 | No | ||
| 185 | RORC | 1740121 | 18449 | -1.922 | -0.0409 | No | ||
| 186 | BCL6 | 940100 | 18581 | -3.177 | 0.0019 | No |

