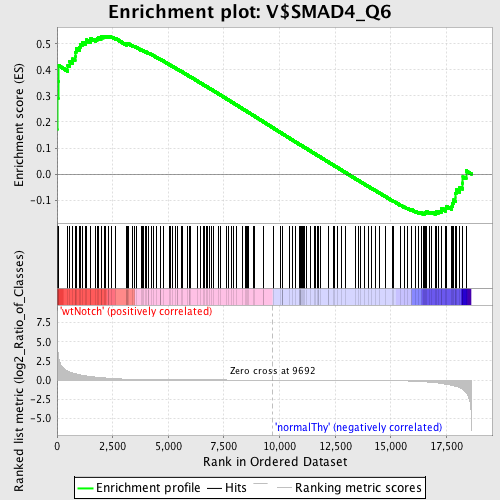
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$SMAD4_Q6 |
| Enrichment Score (ES) | 0.5297601 |
| Normalized Enrichment Score (NES) | 1.2745283 |
| Nominal p-value | 0.049152542 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EEF1A1 | 1980193 1990195 4670735 | 0 | 9.007 | 0.1714 | Yes | ||
| 2 | ARID1A | 2630022 1690551 4810110 | 3 | 6.330 | 0.2917 | Yes | ||
| 3 | MCRS1 | 110121 2340592 5270484 | 42 | 3.506 | 0.3564 | Yes | ||
| 4 | PCBP4 | 6350722 | 49 | 3.258 | 0.4181 | Yes | ||
| 5 | NSBP1 | 2260427 | 483 | 1.143 | 0.4163 | Yes | ||
| 6 | POU2AF1 | 5690242 | 567 | 1.036 | 0.4316 | Yes | ||
| 7 | SFRS6 | 60224 | 699 | 0.926 | 0.4421 | Yes | ||
| 8 | EIF4A1 | 1990341 2810300 | 806 | 0.852 | 0.4526 | Yes | ||
| 9 | ATP2A2 | 1090075 3990279 | 812 | 0.849 | 0.4684 | Yes | ||
| 10 | UBE4B | 780008 3610154 | 852 | 0.812 | 0.4818 | Yes | ||
| 11 | OTX2 | 1190471 | 999 | 0.685 | 0.4869 | Yes | ||
| 12 | HYAL2 | 1500152 | 1034 | 0.665 | 0.4977 | Yes | ||
| 13 | PSMC5 | 2760315 6550021 | 1127 | 0.610 | 0.5043 | Yes | ||
| 14 | HOXB1 | 2120110 | 1273 | 0.548 | 0.5069 | Yes | ||
| 15 | POLR2I | 4060400 | 1307 | 0.535 | 0.5153 | Yes | ||
| 16 | GDF1 | 2350132 | 1495 | 0.458 | 0.5139 | Yes | ||
| 17 | NOTCH3 | 3060603 | 1514 | 0.451 | 0.5215 | Yes | ||
| 18 | FKBP10 | 1340706 | 1733 | 0.381 | 0.5169 | Yes | ||
| 19 | TNFRSF12A | 4810253 | 1794 | 0.364 | 0.5206 | Yes | ||
| 20 | LMO4 | 3800746 | 1841 | 0.351 | 0.5248 | Yes | ||
| 21 | VAPA | 2480594 | 1974 | 0.316 | 0.5237 | Yes | ||
| 22 | UBE2E1 | 4230064 1090440 | 1985 | 0.313 | 0.5291 | Yes | ||
| 23 | RPS6KA3 | 1980707 | 2107 | 0.281 | 0.5279 | Yes | ||
| 24 | CASKIN2 | 2030113 | 2193 | 0.262 | 0.5282 | Yes | ||
| 25 | TBX6 | 4730139 6840647 | 2295 | 0.237 | 0.5273 | Yes | ||
| 26 | TCF4 | 520021 | 2331 | 0.231 | 0.5298 | Yes | ||
| 27 | RPS8 | 4120136 | 2465 | 0.204 | 0.5264 | No | ||
| 28 | PBX3 | 1300424 3710577 6180575 | 2632 | 0.175 | 0.5208 | No | ||
| 29 | PRKAG1 | 6450091 | 3107 | 0.120 | 0.4974 | No | ||
| 30 | TRPS1 | 870100 | 3123 | 0.118 | 0.4988 | No | ||
| 31 | PRKCZ | 3780279 | 3157 | 0.115 | 0.4992 | No | ||
| 32 | RALA | 2680471 | 3168 | 0.114 | 0.5009 | No | ||
| 33 | AP2M1 | 2570338 | 3188 | 0.112 | 0.5020 | No | ||
| 34 | PFN2 | 730040 5700026 | 3403 | 0.092 | 0.4921 | No | ||
| 35 | MMP11 | 1980619 | 3465 | 0.088 | 0.4905 | No | ||
| 36 | AMMECR1 | 3800435 | 3568 | 0.080 | 0.4865 | No | ||
| 37 | PPP3CA | 4760332 6760092 | 3770 | 0.066 | 0.4768 | No | ||
| 38 | SLC22A17 | 540452 4200253 | 3841 | 0.062 | 0.4742 | No | ||
| 39 | SSBP3 | 870717 1050377 7040551 | 3872 | 0.061 | 0.4738 | No | ||
| 40 | THRA | 6770341 110164 1990600 | 3973 | 0.056 | 0.4694 | No | ||
| 41 | NR2F1 | 4120528 360402 | 3999 | 0.054 | 0.4691 | No | ||
| 42 | TMEM25 | 2470575 2570400 | 4098 | 0.050 | 0.4647 | No | ||
| 43 | FASTK | 2690053 4280288 4610551 6370292 | 4128 | 0.049 | 0.4641 | No | ||
| 44 | DHX40 | 360672 | 4232 | 0.045 | 0.4594 | No | ||
| 45 | KCTD15 | 2810735 | 4331 | 0.042 | 0.4549 | No | ||
| 46 | COL1A2 | 380364 | 4477 | 0.038 | 0.4477 | No | ||
| 47 | ACACA | 2490612 2680369 | 4648 | 0.034 | 0.4392 | No | ||
| 48 | COL25A1 | 1580072 4280537 4480168 5290441 7040563 | 4658 | 0.034 | 0.4393 | No | ||
| 49 | HOXA9 | 610494 4730040 5130601 | 4660 | 0.034 | 0.4399 | No | ||
| 50 | AVP | 2100113 | 4766 | 0.032 | 0.4348 | No | ||
| 51 | PDGFRA | 2940332 | 5037 | 0.027 | 0.4207 | No | ||
| 52 | ARHGEF19 | 940102 | 5080 | 0.026 | 0.4189 | No | ||
| 53 | NR1D1 | 2360471 770746 6590204 | 5178 | 0.025 | 0.4141 | No | ||
| 54 | KIRREL3 | 2630195 | 5327 | 0.023 | 0.4066 | No | ||
| 55 | TRIM3 | 5570156 | 5397 | 0.022 | 0.4032 | No | ||
| 56 | HOXA3 | 3610632 6020358 | 5425 | 0.022 | 0.4022 | No | ||
| 57 | WNT1 | 4780148 | 5605 | 0.020 | 0.3929 | No | ||
| 58 | CREB1 | 1500717 2230358 3610600 6550601 | 5622 | 0.020 | 0.3924 | No | ||
| 59 | TENC1 | 1980403 7050575 | 5846 | 0.017 | 0.3806 | No | ||
| 60 | CLOCK | 1410070 2940292 3390075 | 5935 | 0.017 | 0.3762 | No | ||
| 61 | SULF1 | 430575 | 5945 | 0.017 | 0.3760 | No | ||
| 62 | LAMC2 | 450692 2680041 4200059 4670148 | 6011 | 0.016 | 0.3728 | No | ||
| 63 | DACH2 | 460605 1500253 | 6314 | 0.014 | 0.3567 | No | ||
| 64 | HOXA10 | 6110397 7100458 | 6442 | 0.013 | 0.3501 | No | ||
| 65 | NXPH3 | 5860292 | 6575 | 0.012 | 0.3432 | No | ||
| 66 | SP8 | 4060576 | 6627 | 0.012 | 0.3406 | No | ||
| 67 | PHF8 | 70722 | 6718 | 0.011 | 0.3360 | No | ||
| 68 | LMO1 | 3170528 | 6765 | 0.011 | 0.3337 | No | ||
| 69 | ADAMTS4 | 430435 | 6829 | 0.011 | 0.3305 | No | ||
| 70 | NTF5 | 6110239 | 6955 | 0.010 | 0.3239 | No | ||
| 71 | PURA | 3870156 | 7035 | 0.010 | 0.3198 | No | ||
| 72 | IL17RC | 630164 2810593 | 7270 | 0.008 | 0.3073 | No | ||
| 73 | FGF4 | 6550528 | 7333 | 0.008 | 0.3041 | No | ||
| 74 | DOK1 | 1050279 2680112 5220273 | 7620 | 0.007 | 0.2887 | No | ||
| 75 | SLC12A5 | 1980692 | 7708 | 0.007 | 0.2841 | No | ||
| 76 | WNT3 | 2360685 | 7844 | 0.006 | 0.2769 | No | ||
| 77 | AKAP4 | 2190731 3520619 | 7937 | 0.006 | 0.2720 | No | ||
| 78 | SST | 6590142 | 8056 | 0.005 | 0.2657 | No | ||
| 79 | POU4F3 | 2690035 | 8313 | 0.004 | 0.2520 | No | ||
| 80 | SDR-O | 840273 780592 5570537 5900309 | 8450 | 0.004 | 0.2447 | No | ||
| 81 | IL23A | 6290044 | 8510 | 0.004 | 0.2415 | No | ||
| 82 | CALD1 | 1770129 1940397 | 8553 | 0.004 | 0.2393 | No | ||
| 83 | ADORA2B | 6860253 | 8604 | 0.003 | 0.2367 | No | ||
| 84 | LGI1 | 1850020 | 8832 | 0.003 | 0.2244 | No | ||
| 85 | CALM1 | 380128 | 8853 | 0.003 | 0.2234 | No | ||
| 86 | CHRM1 | 4280619 | 9256 | 0.001 | 0.2017 | No | ||
| 87 | CXADR | 6370068 | 9275 | 0.001 | 0.2007 | No | ||
| 88 | CACNG3 | 520093 | 9732 | -0.000 | 0.1760 | No | ||
| 89 | ATP6V1E2 | 6840706 | 10028 | -0.001 | 0.1600 | No | ||
| 90 | SUV39H2 | 1450672 6370242 | 10128 | -0.001 | 0.1547 | No | ||
| 91 | PTBP2 | 1240398 | 10138 | -0.001 | 0.1542 | No | ||
| 92 | RHOQ | 520161 | 10146 | -0.001 | 0.1539 | No | ||
| 93 | PAK3 | 4210136 | 10424 | -0.002 | 0.1389 | No | ||
| 94 | LASS1 | 3440746 6110152 | 10563 | -0.003 | 0.1315 | No | ||
| 95 | CACNA1G | 3060161 4810706 5550722 | 10720 | -0.003 | 0.1231 | No | ||
| 96 | ABCA1 | 6290156 | 10872 | -0.004 | 0.1150 | No | ||
| 97 | EPHB1 | 4610056 | 10898 | -0.004 | 0.1137 | No | ||
| 98 | EVX1 | 1980471 | 10932 | -0.004 | 0.1120 | No | ||
| 99 | CDH13 | 5130368 5340114 | 10944 | -0.004 | 0.1115 | No | ||
| 100 | LOXL3 | 4010168 | 10949 | -0.004 | 0.1113 | No | ||
| 101 | MAFB | 1230471 | 11001 | -0.004 | 0.1086 | No | ||
| 102 | FGF16 | 4210440 | 11043 | -0.004 | 0.1065 | No | ||
| 103 | FOXI1 | 3710095 6620195 | 11052 | -0.004 | 0.1061 | No | ||
| 104 | ADAMTSL1 | 2970092 | 11115 | -0.004 | 0.1029 | No | ||
| 105 | NTRK3 | 4050687 4070167 4610026 6100484 | 11196 | -0.005 | 0.0986 | No | ||
| 106 | FLRT1 | 4540348 | 11390 | -0.005 | 0.0883 | No | ||
| 107 | WDR13 | 3840520 4670064 | 11573 | -0.006 | 0.0785 | No | ||
| 108 | LHB | 1450368 | 11632 | -0.006 | 0.0755 | No | ||
| 109 | NFATC4 | 2470735 | 11722 | -0.007 | 0.0708 | No | ||
| 110 | DLGAP4 | 4570672 5690161 | 11737 | -0.007 | 0.0702 | No | ||
| 111 | TBX5 | 2900132 6370484 | 11744 | -0.007 | 0.0700 | No | ||
| 112 | DSCR1 | 4540048 6220039 | 11835 | -0.007 | 0.0652 | No | ||
| 113 | IRS1 | 1190204 | 12178 | -0.009 | 0.0469 | No | ||
| 114 | BNC2 | 4810603 | 12209 | -0.009 | 0.0454 | No | ||
| 115 | MEF2C | 670025 780338 | 12418 | -0.010 | 0.0343 | No | ||
| 116 | PLEC1 | 6520292 50500 380075 1570088 2510692 2630497 2680017 2900746 4560497 4850376 6290398 7040471 | 12482 | -0.010 | 0.0311 | No | ||
| 117 | AGER | 5860538 5910180 | 12580 | -0.011 | 0.0261 | No | ||
| 118 | DHRS3 | 360609 | 12770 | -0.012 | 0.0160 | No | ||
| 119 | PLA2G2F | 3190646 | 12954 | -0.013 | 0.0064 | No | ||
| 120 | EPHA2 | 5890056 | 13415 | -0.017 | -0.0182 | No | ||
| 121 | KCNN2 | 990025 3520524 | 13556 | -0.019 | -0.0255 | No | ||
| 122 | LHX4 | 6940551 | 13625 | -0.020 | -0.0288 | No | ||
| 123 | HOXA6 | 2340333 | 13812 | -0.022 | -0.0384 | No | ||
| 124 | ETV4 | 2450605 | 13983 | -0.025 | -0.0472 | No | ||
| 125 | PPM2C | 3360209 | 14148 | -0.028 | -0.0555 | No | ||
| 126 | FMO2 | 3850338 | 14303 | -0.031 | -0.0633 | No | ||
| 127 | RGS3 | 60670 540736 1340180 1500369 3390735 4010131 4610402 6380114 | 14506 | -0.035 | -0.0735 | No | ||
| 128 | MYLK | 4010600 7000364 | 14743 | -0.043 | -0.0855 | No | ||
| 129 | FOXB1 | 4920270 5290463 | 15059 | -0.058 | -0.1015 | No | ||
| 130 | CDK6 | 4920253 | 15139 | -0.062 | -0.1046 | No | ||
| 131 | APBB1 | 2690338 | 15454 | -0.086 | -0.1199 | No | ||
| 132 | HNRPUL1 | 2120008 4920408 5570180 6370609 6770403 | 15608 | -0.101 | -0.1263 | No | ||
| 133 | GNB2 | 2350053 | 15749 | -0.115 | -0.1317 | No | ||
| 134 | RORA | 380164 5220215 | 15921 | -0.137 | -0.1384 | No | ||
| 135 | ELOVL5 | 3800170 | 15946 | -0.140 | -0.1370 | No | ||
| 136 | VDR | 130156 510438 | 16112 | -0.164 | -0.1428 | No | ||
| 137 | HNRPK | 1190348 2060411 2570092 6840551 | 16256 | -0.185 | -0.1471 | No | ||
| 138 | CRK | 1230162 4780128 | 16356 | -0.203 | -0.1486 | No | ||
| 139 | SIPA1 | 5220687 | 16478 | -0.225 | -0.1509 | No | ||
| 140 | HHEX | 2340575 | 16533 | -0.235 | -0.1493 | No | ||
| 141 | USP2 | 1190292 1240253 4850035 | 16553 | -0.238 | -0.1458 | No | ||
| 142 | PPP3CB | 6020156 | 16598 | -0.248 | -0.1435 | No | ||
| 143 | FLI1 | 3990142 | 16746 | -0.273 | -0.1462 | No | ||
| 144 | NFKB2 | 2320670 | 16848 | -0.300 | -0.1460 | No | ||
| 145 | YPEL1 | 6590435 | 16994 | -0.337 | -0.1474 | No | ||
| 146 | NAB2 | 1570014 | 17056 | -0.357 | -0.1439 | No | ||
| 147 | LDB1 | 5270601 | 17164 | -0.395 | -0.1422 | No | ||
| 148 | SLC25A14 | 4760180 | 17263 | -0.438 | -0.1392 | No | ||
| 149 | SNX12 | 4060026 | 17266 | -0.439 | -0.1309 | No | ||
| 150 | TRIM8 | 6130441 | 17475 | -0.533 | -0.1321 | No | ||
| 151 | POLD4 | 2320082 | 17512 | -0.550 | -0.1236 | No | ||
| 152 | PHF12 | 870400 3130687 | 17745 | -0.678 | -0.1232 | No | ||
| 153 | DCTN1 | 1450056 6590022 | 17780 | -0.703 | -0.1117 | No | ||
| 154 | PAX1 | 3360253 | 17805 | -0.722 | -0.0993 | No | ||
| 155 | GBA2 | 2900372 | 17912 | -0.795 | -0.0899 | No | ||
| 156 | WASF2 | 4850592 | 17913 | -0.797 | -0.0747 | No | ||
| 157 | DIABLO | 2360170 | 17935 | -0.815 | -0.0604 | No | ||
| 158 | FBS1 | 2570520 | 18100 | -1.002 | -0.0502 | No | ||
| 159 | KLF7 | 50390 | 18236 | -1.223 | -0.0342 | No | ||
| 160 | TRAF3 | 5690647 | 18242 | -1.233 | -0.0110 | No | ||
| 161 | EPC1 | 1570050 5290095 6900193 | 18382 | -1.642 | 0.0127 | No |

