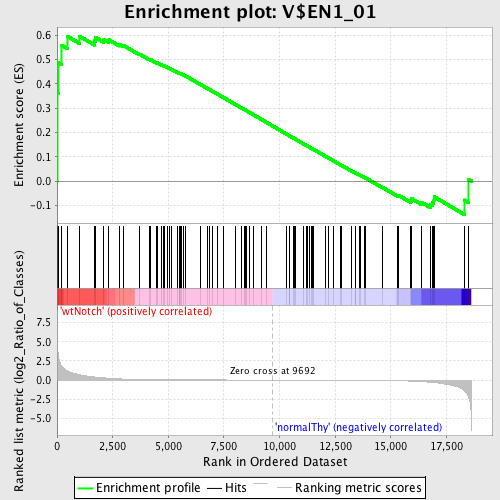
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$EN1_01 |
| Enrichment Score (ES) | 0.5972139 |
| Normalized Enrichment Score (NES) | 1.3311492 |
| Nominal p-value | 0.050167225 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL38 | 70040 4060138 6200601 | 2 | 8.396 | 0.3639 | Yes | ||
| 2 | FIBCD1 | 1500500 | 69 | 2.926 | 0.4871 | Yes | ||
| 3 | SLC6A9 | 2470520 | 204 | 1.842 | 0.5597 | Yes | ||
| 4 | NOTCH1 | 3390114 | 463 | 1.166 | 0.5964 | Yes | ||
| 5 | OTX2 | 1190471 | 999 | 0.685 | 0.5972 | Yes | ||
| 6 | SIPA1L1 | 50292 | 1684 | 0.396 | 0.5775 | No | ||
| 7 | MORF4L2 | 6450133 | 1725 | 0.384 | 0.5919 | No | ||
| 8 | SAV1 | 5420541 | 2103 | 0.282 | 0.5838 | No | ||
| 9 | TCF4 | 520021 | 2331 | 0.231 | 0.5816 | No | ||
| 10 | ZADH2 | 520438 | 2797 | 0.153 | 0.5631 | No | ||
| 11 | CDC14A | 4050132 | 2962 | 0.133 | 0.5600 | No | ||
| 12 | RBM4 | 2360113 5080750 | 3707 | 0.070 | 0.5229 | No | ||
| 13 | NOS1 | 5860129 | 4132 | 0.048 | 0.5021 | No | ||
| 14 | A2BP1 | 2370390 4590593 5550014 | 4218 | 0.046 | 0.4995 | No | ||
| 15 | ELN | 5080347 | 4480 | 0.038 | 0.4871 | No | ||
| 16 | FOXP2 | 3520561 4150372 4760524 | 4495 | 0.038 | 0.4880 | No | ||
| 17 | NEO1 | 1990154 | 4699 | 0.033 | 0.4785 | No | ||
| 18 | TFEB | 3390121 | 4782 | 0.031 | 0.4754 | No | ||
| 19 | TTC12 | 4480632 | 4830 | 0.030 | 0.4742 | No | ||
| 20 | GRIA1 | 1340152 3780750 4920440 | 4957 | 0.028 | 0.4686 | No | ||
| 21 | PDGFRA | 2940332 | 5037 | 0.027 | 0.4655 | No | ||
| 22 | OTP | 2630070 | 5153 | 0.025 | 0.4604 | No | ||
| 23 | RGS6 | 6040601 | 5392 | 0.022 | 0.4485 | No | ||
| 24 | PLCB1 | 2570050 7100102 | 5484 | 0.021 | 0.4445 | No | ||
| 25 | FGF17 | 3130022 | 5518 | 0.021 | 0.4437 | No | ||
| 26 | RAB2B | 6200091 | 5544 | 0.021 | 0.4432 | No | ||
| 27 | RAPGEF5 | 1500603 3450427 6130056 | 5576 | 0.020 | 0.4424 | No | ||
| 28 | GPC3 | 6180735 | 5671 | 0.019 | 0.4382 | No | ||
| 29 | RTKN | 5910253 6380082 | 5765 | 0.018 | 0.4340 | No | ||
| 30 | HOXB8 | 3710450 | 6441 | 0.013 | 0.3981 | No | ||
| 31 | LMO1 | 3170528 | 6765 | 0.011 | 0.3811 | No | ||
| 32 | GPC4 | 5910408 | 6842 | 0.010 | 0.3775 | No | ||
| 33 | UBE2H | 1980142 2970079 | 7004 | 0.010 | 0.3692 | No | ||
| 34 | SYNJ2BP | 4150672 6770706 | 7223 | 0.009 | 0.3579 | No | ||
| 35 | ARHGAP6 | 2060121 | 7460 | 0.008 | 0.3455 | No | ||
| 36 | BDNF | 2940128 3520368 | 8004 | 0.006 | 0.3164 | No | ||
| 37 | IRX4 | 1980102 | 8269 | 0.005 | 0.3024 | No | ||
| 38 | NPR3 | 1580239 | 8419 | 0.004 | 0.2945 | No | ||
| 39 | NRAS | 6900577 | 8451 | 0.004 | 0.2930 | No | ||
| 40 | EFEMP1 | 5860324 | 8496 | 0.004 | 0.2908 | No | ||
| 41 | HOXD10 | 6100039 | 8634 | 0.003 | 0.2835 | No | ||
| 42 | NKX2-2 | 4150731 | 8820 | 0.003 | 0.2737 | No | ||
| 43 | UTY | 5890441 | 9197 | 0.001 | 0.2535 | No | ||
| 44 | GAP43 | 3130504 | 9391 | 0.001 | 0.2431 | No | ||
| 45 | HOXD3 | 6450154 | 10326 | -0.002 | 0.1928 | No | ||
| 46 | HTR7 | 2680176 | 10453 | -0.002 | 0.1861 | No | ||
| 47 | PXMP3 | 2100088 1400458 6650671 | 10456 | -0.002 | 0.1861 | No | ||
| 48 | ATP9B | 3710494 6020239 | 10623 | -0.003 | 0.1772 | No | ||
| 49 | NRG1 | 1050332 | 10677 | -0.003 | 0.1745 | No | ||
| 50 | EYA4 | 5360286 | 10723 | -0.003 | 0.1722 | No | ||
| 51 | PAMCI | 2320687 | 11070 | -0.004 | 0.1537 | No | ||
| 52 | HOXA11 | 6980133 | 11187 | -0.005 | 0.1477 | No | ||
| 53 | ANK2 | 6510546 | 11257 | -0.005 | 0.1442 | No | ||
| 54 | NR4A3 | 2900021 5860095 5910039 | 11354 | -0.005 | 0.1392 | No | ||
| 55 | BCOR | 1660452 3940053 | 11421 | -0.006 | 0.1359 | No | ||
| 56 | MITF | 380056 | 11498 | -0.006 | 0.1320 | No | ||
| 57 | PAX6 | 1190025 | 11545 | -0.006 | 0.1298 | No | ||
| 58 | RARB | 430139 1410138 | 12049 | -0.008 | 0.1030 | No | ||
| 59 | GREM1 | 3940180 | 12219 | -0.009 | 0.0943 | No | ||
| 60 | SLC6A14 | 380368 | 12430 | -0.010 | 0.0834 | No | ||
| 61 | GNRH1 | 430167 | 12751 | -0.012 | 0.0667 | No | ||
| 62 | KLF15 | 4230164 | 12771 | -0.012 | 0.0661 | No | ||
| 63 | MYOG | 3190672 | 13217 | -0.015 | 0.0428 | No | ||
| 64 | ISL1 | 4810239 | 13418 | -0.017 | 0.0328 | No | ||
| 65 | ERBB4 | 4610400 4810017 | 13431 | -0.017 | 0.0329 | No | ||
| 66 | SH3BGRL2 | 1780239 | 13606 | -0.019 | 0.0243 | No | ||
| 67 | PCDH9 | 2100136 | 13631 | -0.020 | 0.0239 | No | ||
| 68 | MAP3K5 | 6020041 6380162 | 13797 | -0.022 | 0.0159 | No | ||
| 69 | LAPTM4B | 2060377 4150398 | 13879 | -0.023 | 0.0126 | No | ||
| 70 | CDX2 | 3850349 | 14621 | -0.039 | -0.0257 | No | ||
| 71 | FHL1 | 60278 | 15297 | -0.073 | -0.0590 | No | ||
| 72 | HOXA4 | 940152 | 15336 | -0.075 | -0.0578 | No | ||
| 73 | EOMES | 2100707 | 15884 | -0.131 | -0.0816 | No | ||
| 74 | CAMLG | 1410184 | 15920 | -0.137 | -0.0776 | No | ||
| 75 | UTX | 2900017 6770452 | 15934 | -0.139 | -0.0723 | No | ||
| 76 | CREM | 840156 6380438 6660041 6660168 | 16364 | -0.204 | -0.0866 | No | ||
| 77 | CTCF | 5340017 | 16777 | -0.280 | -0.0967 | No | ||
| 78 | MAP4K4 | 2360059 | 16877 | -0.307 | -0.0887 | No | ||
| 79 | MAPK14 | 5290731 | 16936 | -0.320 | -0.0780 | No | ||
| 80 | NXF1 | 1570377 | 16945 | -0.323 | -0.0644 | No | ||
| 81 | RDH10 | 6650070 | 18302 | -1.378 | -0.0778 | No | ||
| 82 | MMP14 | 670079 | 18487 | -2.185 | 0.0070 | No |

