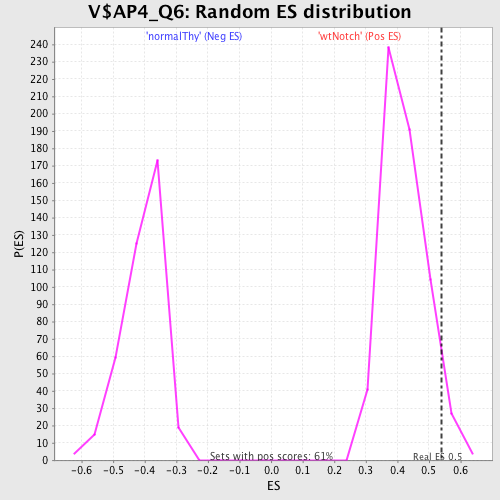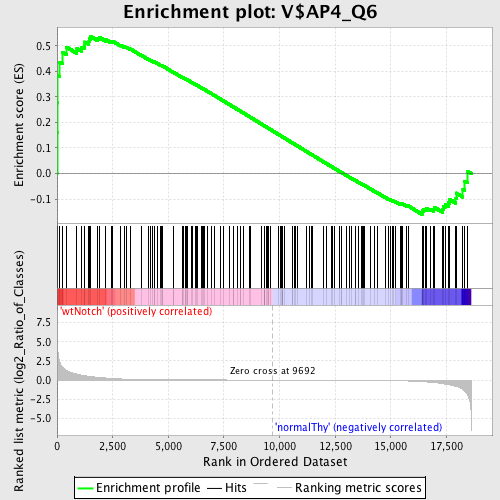
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | wtNotch |
| GeneSet | V$AP4_Q6 |
| Enrichment Score (ES) | 0.5385428 |
| Normalized Enrichment Score (NES) | 1.2803847 |
| Nominal p-value | 0.05123967 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 2630022 1690551 4810110 | 3 | 6.330 | 0.1598 | Yes | ||
| 2 | TPM3 | 670600 3170296 5670167 6650471 | 19 | 4.650 | 0.2766 | Yes | ||
| 3 | INPPL1 | 110717 3120164 | 24 | 4.268 | 0.3842 | Yes | ||
| 4 | UBTF | 4210450 6840403 | 121 | 2.292 | 0.4369 | Yes | ||
| 5 | PTPRS | 510414 4010215 6400139 6770348 | 235 | 1.743 | 0.4749 | Yes | ||
| 6 | SYTL1 | 510487 | 427 | 1.224 | 0.4955 | Yes | ||
| 7 | PRKACA | 2640731 4050048 | 889 | 0.782 | 0.4903 | Yes | ||
| 8 | PRICKLE1 | 450671 | 1085 | 0.637 | 0.4958 | Yes | ||
| 9 | PTK7 | 380315 | 1222 | 0.569 | 0.5029 | Yes | ||
| 10 | HMGN2 | 3140091 | 1250 | 0.558 | 0.5155 | Yes | ||
| 11 | RAB3A | 3940288 | 1429 | 0.488 | 0.5182 | Yes | ||
| 12 | ABHD4 | 6760450 | 1457 | 0.475 | 0.5288 | Yes | ||
| 13 | TRAPPC3 | 2030176 6020609 6590373 6760019 | 1492 | 0.460 | 0.5385 | Yes | ||
| 14 | VAT1 | 1050040 | 1827 | 0.355 | 0.5294 | No | ||
| 15 | NDST2 | 6290711 | 1911 | 0.330 | 0.5333 | No | ||
| 16 | TRAF4 | 3060041 4920528 6980286 | 2176 | 0.267 | 0.5257 | No | ||
| 17 | VAMP3 | 7100050 | 2425 | 0.212 | 0.5177 | No | ||
| 18 | BSCL2 | 2680671 | 2495 | 0.199 | 0.5190 | No | ||
| 19 | RAMP2 | 6860037 | 2851 | 0.146 | 0.5034 | No | ||
| 20 | STOML2 | 1940487 | 3023 | 0.128 | 0.4974 | No | ||
| 21 | CNNM2 | 2650100 | 3124 | 0.118 | 0.4950 | No | ||
| 22 | GAS7 | 2120358 6450494 | 3298 | 0.102 | 0.4882 | No | ||
| 23 | TNFSF4 | 3940594 | 3805 | 0.064 | 0.4624 | No | ||
| 24 | CREB3L1 | 2350687 | 4097 | 0.050 | 0.4479 | No | ||
| 25 | GRIK3 | 6380592 | 4197 | 0.046 | 0.4437 | No | ||
| 26 | EPOR | 5290435 | 4271 | 0.044 | 0.4409 | No | ||
| 27 | SOX12 | 1780037 | 4369 | 0.041 | 0.4367 | No | ||
| 28 | RPS6KB1 | 1450427 5080110 6200563 | 4372 | 0.041 | 0.4376 | No | ||
| 29 | INHA | 6100102 | 4527 | 0.037 | 0.4302 | No | ||
| 30 | TMOD3 | 840088 | 4664 | 0.034 | 0.4237 | No | ||
| 31 | ERN1 | 2360403 | 4692 | 0.033 | 0.4231 | No | ||
| 32 | ZRANB1 | 510373 4540278 | 4738 | 0.032 | 0.4215 | No | ||
| 33 | FGF8 | 6350504 | 4756 | 0.032 | 0.4213 | No | ||
| 34 | FRAS1 | 5080170 5670347 | 5236 | 0.024 | 0.3960 | No | ||
| 35 | HAPLN2 | 2900647 | 5237 | 0.024 | 0.3966 | No | ||
| 36 | CTDSP1 | 6650215 6940731 | 5626 | 0.020 | 0.3761 | No | ||
| 37 | PALM2 | 4120068 7100398 | 5628 | 0.020 | 0.3766 | No | ||
| 38 | PTCH2 | 2900092 | 5694 | 0.019 | 0.3735 | No | ||
| 39 | HOXC4 | 1940193 | 5751 | 0.018 | 0.3710 | No | ||
| 40 | RTKN | 5910253 6380082 | 5765 | 0.018 | 0.3707 | No | ||
| 41 | PITX3 | 3130162 | 5818 | 0.018 | 0.3684 | No | ||
| 42 | CACNB2 | 1500095 7330707 | 5868 | 0.017 | 0.3662 | No | ||
| 43 | PLXNB3 | 130008 | 5881 | 0.017 | 0.3659 | No | ||
| 44 | KCNE1L | 4670092 | 6052 | 0.016 | 0.3571 | No | ||
| 45 | ANXA8 | 4780022 | 6064 | 0.016 | 0.3569 | No | ||
| 46 | DLL3 | 4010093 | 6209 | 0.015 | 0.3495 | No | ||
| 47 | TNNT2 | 2450364 | 6276 | 0.014 | 0.3463 | No | ||
| 48 | DOCK4 | 5910102 | 6324 | 0.014 | 0.3441 | No | ||
| 49 | PICALM | 1940280 | 6478 | 0.013 | 0.3361 | No | ||
| 50 | EMILIN3 | 5130181 | 6544 | 0.012 | 0.3329 | No | ||
| 51 | NXPH3 | 5860292 | 6575 | 0.012 | 0.3316 | No | ||
| 52 | SYTL2 | 580097 5390576 6770603 | 6613 | 0.012 | 0.3299 | No | ||
| 53 | ABLIM1 | 580170 3710338 6520504 | 6741 | 0.011 | 0.3233 | No | ||
| 54 | NEUROG3 | 6980451 | 6935 | 0.010 | 0.3131 | No | ||
| 55 | GRIN2B | 3800333 | 7052 | 0.010 | 0.3071 | No | ||
| 56 | EYA1 | 1450278 5220390 | 7336 | 0.008 | 0.2920 | No | ||
| 57 | OBSCN | 3610129 | 7479 | 0.008 | 0.2845 | No | ||
| 58 | ASB16 | 1940152 | 7758 | 0.007 | 0.2696 | No | ||
| 59 | WNT4 | 4150619 | 7931 | 0.006 | 0.2604 | No | ||
| 60 | CHRNG | 5860273 | 7935 | 0.006 | 0.2604 | No | ||
| 61 | HOXC12 | 5270411 | 8110 | 0.005 | 0.2511 | No | ||
| 62 | TRPC4 | 6040022 | 8233 | 0.005 | 0.2446 | No | ||
| 63 | TNRC4 | 4050156 | 8253 | 0.005 | 0.2437 | No | ||
| 64 | STARD13 | 540722 | 8398 | 0.004 | 0.2360 | No | ||
| 65 | MID2 | 4120309 | 8630 | 0.003 | 0.2236 | No | ||
| 66 | FBXO24 | 1230132 4610121 | 8707 | 0.003 | 0.2196 | No | ||
| 67 | PRKCQ | 2260170 3870193 | 9203 | 0.001 | 0.1928 | No | ||
| 68 | ASB5 | 2760575 | 9299 | 0.001 | 0.1877 | No | ||
| 69 | LBX1 | 1300164 4060204 | 9304 | 0.001 | 0.1875 | No | ||
| 70 | ELF4 | 4850193 | 9326 | 0.001 | 0.1864 | No | ||
| 71 | GNB3 | 1230167 | 9402 | 0.001 | 0.1824 | No | ||
| 72 | KCNMA1 | 1450402 | 9448 | 0.001 | 0.1800 | No | ||
| 73 | PCDH12 | 5340044 | 9519 | 0.001 | 0.1762 | No | ||
| 74 | SPOCK2 | 6650369 | 9522 | 0.001 | 0.1761 | No | ||
| 75 | FOSL2 | 870044 1230427 6520433 | 9576 | 0.000 | 0.1732 | No | ||
| 76 | MAP3K13 | 3190017 | 9941 | -0.001 | 0.1536 | No | ||
| 77 | SOST | 1170195 | 10029 | -0.001 | 0.1489 | No | ||
| 78 | GNG3 | 6980397 | 10097 | -0.001 | 0.1453 | No | ||
| 79 | MYBPH | 2190711 | 10110 | -0.001 | 0.1447 | No | ||
| 80 | SUV39H2 | 1450672 6370242 | 10128 | -0.001 | 0.1438 | No | ||
| 81 | FGF12 | 1740446 2360037 | 10144 | -0.001 | 0.1430 | No | ||
| 82 | PCYT1B | 1780402 | 10241 | -0.002 | 0.1378 | No | ||
| 83 | CADPS | 2320181 | 10562 | -0.003 | 0.1206 | No | ||
| 84 | NES | 1190672 2450167 | 10581 | -0.003 | 0.1197 | No | ||
| 85 | KCNQ4 | 3800438 7050400 | 10648 | -0.003 | 0.1162 | No | ||
| 86 | CACNA1G | 3060161 4810706 5550722 | 10720 | -0.003 | 0.1124 | No | ||
| 87 | PGF | 4810593 | 10786 | -0.003 | 0.1090 | No | ||
| 88 | RGS7 | 3390044 | 11203 | -0.005 | 0.0866 | No | ||
| 89 | FOXD3 | 6550156 | 11353 | -0.005 | 0.0786 | No | ||
| 90 | PROK2 | 540538 | 11419 | -0.006 | 0.0753 | No | ||
| 91 | XPNPEP1 | 2640750 3850315 6200242 | 11493 | -0.006 | 0.0715 | No | ||
| 92 | KIRREL2 | 130168 3440097 | 11987 | -0.008 | 0.0450 | No | ||
| 93 | RBMS3 | 6110722 | 12095 | -0.008 | 0.0394 | No | ||
| 94 | RXRG | 7040008 | 12323 | -0.009 | 0.0273 | No | ||
| 95 | JMJD1C | 940575 2120025 | 12357 | -0.010 | 0.0258 | No | ||
| 96 | HOXB5 | 2900538 | 12487 | -0.010 | 0.0191 | No | ||
| 97 | CPEB3 | 3940164 | 12700 | -0.011 | 0.0079 | No | ||
| 98 | DHRS3 | 360609 | 12770 | -0.012 | 0.0045 | No | ||
| 99 | LOXL4 | 6520095 | 12795 | -0.012 | 0.0035 | No | ||
| 100 | MLLT7 | 4480707 | 13009 | -0.014 | -0.0077 | No | ||
| 101 | FHL3 | 1090369 2650017 | 13142 | -0.015 | -0.0145 | No | ||
| 102 | WNT9A | 3830711 | 13234 | -0.015 | -0.0190 | No | ||
| 103 | EPHA2 | 5890056 | 13415 | -0.017 | -0.0283 | No | ||
| 104 | ARHGEF2 | 3360577 | 13426 | -0.017 | -0.0284 | No | ||
| 105 | KCNN2 | 990025 3520524 | 13556 | -0.019 | -0.0350 | No | ||
| 106 | PABPC5 | 2320408 | 13684 | -0.020 | -0.0413 | No | ||
| 107 | FBXO40 | 2190286 5670253 | 13714 | -0.021 | -0.0424 | No | ||
| 108 | NPPA | 6550328 | 13790 | -0.022 | -0.0459 | No | ||
| 109 | HOXA6 | 2340333 | 13812 | -0.022 | -0.0464 | No | ||
| 110 | AKAP2 | 5550347 | 14090 | -0.027 | -0.0608 | No | ||
| 111 | HFE2 | 2100138 3850148 | 14271 | -0.030 | -0.0697 | No | ||
| 112 | ARNT | 1170672 5670711 | 14409 | -0.033 | -0.0763 | No | ||
| 113 | MYLK | 4010600 7000364 | 14743 | -0.043 | -0.0932 | No | ||
| 114 | PDE3A | 380068 | 14901 | -0.050 | -0.1005 | No | ||
| 115 | RASGRF2 | 1990047 | 14966 | -0.053 | -0.1026 | No | ||
| 116 | AICDA | 2340487 3800672 | 15077 | -0.058 | -0.1071 | No | ||
| 117 | WNT2B | 3130088 5960575 | 15115 | -0.061 | -0.1075 | No | ||
| 118 | BCL2L2 | 2760692 6770739 | 15194 | -0.066 | -0.1101 | No | ||
| 119 | NANOS1 | 1300402 | 15425 | -0.083 | -0.1204 | No | ||
| 120 | TCF7 | 3800736 5390181 | 15467 | -0.088 | -0.1204 | No | ||
| 121 | WDTC1 | 1690541 | 15489 | -0.090 | -0.1193 | No | ||
| 122 | ZNF385 | 450176 4810358 | 15514 | -0.092 | -0.1183 | No | ||
| 123 | SCAMP5 | 5550368 6290021 | 15724 | -0.112 | -0.1268 | No | ||
| 124 | FMO5 | 4810725 5390086 | 15777 | -0.119 | -0.1266 | No | ||
| 125 | ESRRA | 6550242 | 16407 | -0.211 | -0.1553 | No | ||
| 126 | LAT | 3170025 | 16412 | -0.213 | -0.1501 | No | ||
| 127 | EYA3 | 1580195 6510632 | 16438 | -0.218 | -0.1460 | No | ||
| 128 | AMPH | 2260338 3140576 | 16468 | -0.223 | -0.1419 | No | ||
| 129 | TMOD4 | 2360021 4590722 5910520 | 16540 | -0.236 | -0.1397 | No | ||
| 130 | COL7A1 | 1230168 2190072 | 16613 | -0.250 | -0.1373 | No | ||
| 131 | ADRBK1 | 1340333 | 16790 | -0.283 | -0.1397 | No | ||
| 132 | MESDC1 | 2030019 | 16939 | -0.321 | -0.1396 | No | ||
| 133 | EMP3 | 1500722 1570039 | 16956 | -0.325 | -0.1323 | No | ||
| 134 | PTPN7 | 3450110 | 17336 | -0.473 | -0.1408 | No | ||
| 135 | LCK | 3360142 | 17374 | -0.493 | -0.1304 | No | ||
| 136 | RAB27A | 450019 | 17437 | -0.517 | -0.1207 | No | ||
| 137 | GATA3 | 6130068 | 17582 | -0.581 | -0.1138 | No | ||
| 138 | BACH1 | 290195 | 17652 | -0.618 | -0.1019 | No | ||
| 139 | RUNX1 | 3840711 | 17925 | -0.807 | -0.0962 | No | ||
| 140 | ARHGAP24 | 540278 4540025 | 17942 | -0.821 | -0.0763 | No | ||
| 141 | KLF13 | 4670563 | 18239 | -1.228 | -0.0613 | No | ||
| 142 | HSD11B1 | 450066 5550408 | 18314 | -1.402 | -0.0299 | No | ||
| 143 | BCL11B | 2680673 | 18435 | -1.828 | 0.0098 | No |