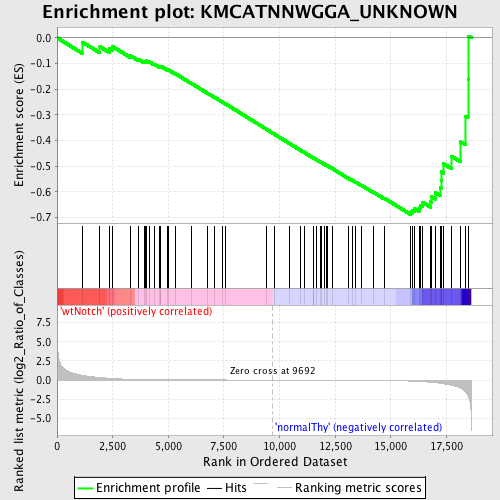
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | KMCATNNWGGA_UNKNOWN |
| Enrichment Score (ES) | -0.68860865 |
| Normalized Enrichment Score (NES) | -1.5246698 |
| Nominal p-value | 0.011337869 |
| FDR q-value | 0.13161798 |
| FWER p-Value | 0.692 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ESM1 | 1410594 | 1143 | 0.604 | -0.0170 | No | ||
| 2 | CABP1 | 6350750 | 1924 | 0.328 | -0.0348 | No | ||
| 3 | ABL1 | 1050593 2030050 4010114 | 2358 | 0.225 | -0.0415 | No | ||
| 4 | CAB39L | 5670040 | 2499 | 0.198 | -0.0344 | No | ||
| 5 | GAS7 | 2120358 6450494 | 3298 | 0.102 | -0.0699 | No | ||
| 6 | TSC1 | 1850672 | 3660 | 0.073 | -0.0840 | No | ||
| 7 | PDE4D | 2470528 6660014 | 3914 | 0.059 | -0.0933 | No | ||
| 8 | CNOT3 | 130519 4480292 1990609 | 3985 | 0.055 | -0.0929 | No | ||
| 9 | NR2F1 | 4120528 360402 | 3999 | 0.054 | -0.0896 | No | ||
| 10 | NR2F2 | 3170609 3310577 | 4149 | 0.048 | -0.0941 | No | ||
| 11 | SOX12 | 1780037 | 4369 | 0.041 | -0.1029 | No | ||
| 12 | ENAH | 1690292 5700300 | 4595 | 0.035 | -0.1124 | No | ||
| 13 | SOCS5 | 3830398 7100093 | 4649 | 0.034 | -0.1127 | No | ||
| 14 | CDC45L | 70537 3130114 | 4662 | 0.034 | -0.1109 | No | ||
| 15 | STAG1 | 1190300 1400722 4590100 | 4962 | 0.028 | -0.1249 | No | ||
| 16 | PNPT1 | 5270433 | 4984 | 0.028 | -0.1240 | No | ||
| 17 | GEMIN4 | 130278 2230075 | 5305 | 0.023 | -0.1395 | No | ||
| 18 | RBM16 | 6290286 6400181 | 6030 | 0.016 | -0.1773 | No | ||
| 19 | LMO1 | 3170528 | 6765 | 0.011 | -0.2161 | No | ||
| 20 | DUSP7 | 2940309 | 7071 | 0.009 | -0.2318 | No | ||
| 21 | AP1G1 | 2360671 | 7414 | 0.008 | -0.2497 | No | ||
| 22 | UFD1L | 870020 | 7546 | 0.007 | -0.2562 | No | ||
| 23 | SNX5 | 1570021 6860609 | 9427 | 0.001 | -0.3574 | No | ||
| 24 | KCNJ9 | 3440292 | 9757 | -0.000 | -0.3751 | No | ||
| 25 | PPP4R2 | 5890035 | 10436 | -0.002 | -0.4115 | No | ||
| 26 | SP3 | 3840338 | 10919 | -0.004 | -0.4372 | No | ||
| 27 | ADAMTSL1 | 2970092 | 11115 | -0.004 | -0.4474 | No | ||
| 28 | OSBPL6 | 3800072 | 11514 | -0.006 | -0.4684 | No | ||
| 29 | PAX6 | 1190025 | 11545 | -0.006 | -0.4696 | No | ||
| 30 | IL2 | 1770725 | 11670 | -0.006 | -0.4758 | No | ||
| 31 | ZBTB11 | 6520092 | 11824 | -0.007 | -0.4835 | No | ||
| 32 | STRN4 | 3440711 | 11865 | -0.007 | -0.4851 | No | ||
| 33 | SDCCAG10 | 2630286 | 11997 | -0.008 | -0.4916 | No | ||
| 34 | CRAT | 540020 2060364 3520148 | 12105 | -0.008 | -0.4968 | No | ||
| 35 | SYNCRIP | 1690195 3140113 4670279 | 12143 | -0.009 | -0.4981 | No | ||
| 36 | CELSR3 | 5270731 7100082 | 12387 | -0.010 | -0.5105 | No | ||
| 37 | CSNK1A1 | 2340427 | 13119 | -0.015 | -0.5488 | No | ||
| 38 | DMD | 1740041 3990332 | 13276 | -0.016 | -0.5560 | No | ||
| 39 | EPHA2 | 5890056 | 13415 | -0.017 | -0.5622 | No | ||
| 40 | SPRED2 | 510138 540195 | 13688 | -0.020 | -0.5753 | No | ||
| 41 | RECK | 840079 | 14217 | -0.029 | -0.6016 | No | ||
| 42 | BCL2L13 | 3170593 | 14705 | -0.042 | -0.6248 | No | ||
| 43 | CRMP1 | 4810014 | 15890 | -0.132 | -0.6789 | Yes | ||
| 44 | ING3 | 110438 2060390 | 15975 | -0.145 | -0.6727 | Yes | ||
| 45 | MAP2K5 | 2030440 | 16064 | -0.156 | -0.6659 | Yes | ||
| 46 | BRUNOL4 | 4490452 5080451 | 16269 | -0.187 | -0.6631 | Yes | ||
| 47 | ANAPC7 | 5260274 | 16340 | -0.198 | -0.6522 | Yes | ||
| 48 | AEBP2 | 2970603 | 16442 | -0.219 | -0.6415 | Yes | ||
| 49 | MAP4K3 | 5390551 | 16794 | -0.284 | -0.6395 | Yes | ||
| 50 | SFRS1 | 2360440 | 16829 | -0.295 | -0.6195 | Yes | ||
| 51 | RSHL2 | 6110014 | 16997 | -0.339 | -0.6035 | Yes | ||
| 52 | FKRP | 130546 | 17216 | -0.420 | -0.5842 | Yes | ||
| 53 | WASL | 3290170 | 17262 | -0.437 | -0.5544 | Yes | ||
| 54 | CGGBP1 | 4810053 | 17283 | -0.445 | -0.5225 | Yes | ||
| 55 | HIC2 | 4010433 | 17372 | -0.492 | -0.4909 | Yes | ||
| 56 | HOXA7 | 5910152 | 17729 | -0.661 | -0.4613 | Yes | ||
| 57 | MRPL24 | 1170014 | 18143 | -1.050 | -0.4060 | Yes | ||
| 58 | ARNTL | 3170463 | 18352 | -1.518 | -0.3051 | Yes | ||
| 59 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 18469 | -2.031 | -0.1613 | Yes | ||
| 60 | DUSP6 | 5910286 7100070 | 18498 | -2.291 | 0.0064 | Yes |

