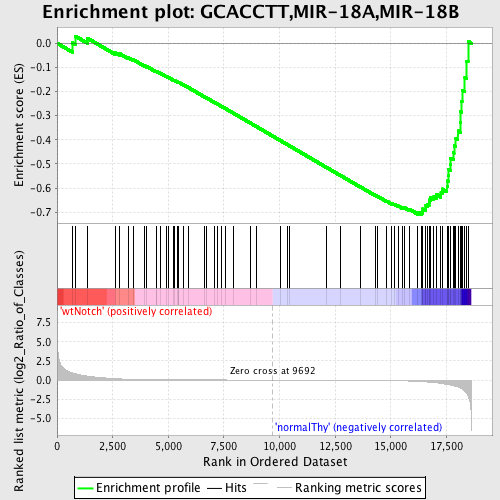
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | GCACCTT,MIR-18A,MIR-18B |
| Enrichment Score (ES) | -0.7092183 |
| Normalized Enrichment Score (NES) | -1.6141268 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09290033 |
| FWER p-Value | 0.247 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDC42 | 1240168 3440278 4480519 5290162 | 682 | 0.936 | 0.0018 | No | ||
| 2 | PHF19 | 3800358 | 816 | 0.845 | 0.0294 | No | ||
| 3 | TEX2 | 1740504 2480102 3060494 | 1373 | 0.508 | 0.0203 | No | ||
| 4 | PARP6 | 3290332 4760167 6900181 | 2610 | 0.179 | -0.0390 | No | ||
| 5 | EHMT1 | 5290433 5910086 6620112 6980014 | 2802 | 0.152 | -0.0430 | No | ||
| 6 | NR3C1 | 630563 | 3215 | 0.110 | -0.0607 | No | ||
| 7 | NFAT5 | 2510411 5890195 6550152 | 3440 | 0.090 | -0.0691 | No | ||
| 8 | PIAS3 | 50204 240739 4060524 | 3949 | 0.057 | -0.0941 | No | ||
| 9 | TARDBP | 870390 2350093 | 4036 | 0.052 | -0.0966 | No | ||
| 10 | ARL15 | 6520692 | 4446 | 0.039 | -0.1171 | No | ||
| 11 | RABGAP1 | 5050397 | 4468 | 0.038 | -0.1166 | No | ||
| 12 | SOCS5 | 3830398 7100093 | 4649 | 0.034 | -0.1249 | No | ||
| 13 | IGF1 | 1990193 3130377 3290280 | 4924 | 0.029 | -0.1385 | No | ||
| 14 | SON | 3120148 5860239 7040403 | 4992 | 0.028 | -0.1410 | No | ||
| 15 | HSF2 | 1400465 | 5232 | 0.024 | -0.1529 | No | ||
| 16 | PRICKLE2 | 780408 | 5263 | 0.024 | -0.1535 | No | ||
| 17 | SH3BP4 | 3140242 | 5432 | 0.022 | -0.1617 | No | ||
| 18 | ESR1 | 4060372 5860193 | 5448 | 0.022 | -0.1616 | No | ||
| 19 | MESP1 | 4280673 | 5464 | 0.021 | -0.1615 | No | ||
| 20 | RAB11FIP2 | 4570040 | 5670 | 0.019 | -0.1718 | No | ||
| 21 | AKR1D1 | 6770026 | 5896 | 0.017 | -0.1832 | No | ||
| 22 | PTGFRN | 4120524 | 6602 | 0.012 | -0.2208 | No | ||
| 23 | CTGF | 4540577 | 6709 | 0.011 | -0.2260 | No | ||
| 24 | ZBTB4 | 6450441 | 7059 | 0.009 | -0.2445 | No | ||
| 25 | ASXL2 | 130670 | 7209 | 0.009 | -0.2521 | No | ||
| 26 | DPP10 | 4730746 | 7407 | 0.008 | -0.2624 | No | ||
| 27 | LIN28 | 6590672 | 7554 | 0.007 | -0.2700 | No | ||
| 28 | BHLHB5 | 6510520 | 7944 | 0.006 | -0.2907 | No | ||
| 29 | BTG3 | 7050079 | 8698 | 0.003 | -0.3312 | No | ||
| 30 | ALCAM | 1050019 | 8965 | 0.002 | -0.3455 | No | ||
| 31 | TRIM2 | 430546 1170286 | 10027 | -0.001 | -0.4027 | No | ||
| 32 | SIM2 | 2100044 | 10364 | -0.002 | -0.4207 | No | ||
| 33 | HIF1A | 5670605 | 10440 | -0.002 | -0.4246 | No | ||
| 34 | GCLC | 2810731 | 12109 | -0.008 | -0.5143 | No | ||
| 35 | MAN1A2 | 5700435 5720114 | 12757 | -0.012 | -0.5487 | No | ||
| 36 | CLASP2 | 2510139 | 13622 | -0.020 | -0.5945 | No | ||
| 37 | NAV1 | 4210519 6620129 | 14317 | -0.032 | -0.6306 | No | ||
| 38 | GLRB | 2510433 4280736 | 14390 | -0.033 | -0.6331 | No | ||
| 39 | TRIOBP | 940750 1300187 3140072 | 14823 | -0.046 | -0.6545 | No | ||
| 40 | MDGA1 | 1170139 | 15050 | -0.057 | -0.6643 | No | ||
| 41 | MEF2D | 5690576 | 15149 | -0.064 | -0.6670 | No | ||
| 42 | CRIM1 | 130491 | 15335 | -0.075 | -0.6739 | No | ||
| 43 | FNBP1 | 6100017 | 15524 | -0.092 | -0.6802 | No | ||
| 44 | CTDSPL | 4670546 | 15622 | -0.102 | -0.6813 | No | ||
| 45 | PURB | 5360138 | 15833 | -0.126 | -0.6874 | No | ||
| 46 | RHOT1 | 6760692 | 16211 | -0.178 | -0.7004 | No | ||
| 47 | SNURF | 3520053 | 16375 | -0.205 | -0.7008 | Yes | ||
| 48 | SAR1A | 3450253 | 16432 | -0.217 | -0.6949 | Yes | ||
| 49 | AEBP2 | 2970603 | 16442 | -0.219 | -0.6863 | Yes | ||
| 50 | ADD3 | 4150739 5390600 | 16573 | -0.243 | -0.6833 | Yes | ||
| 51 | ATXN1 | 5550156 | 16576 | -0.244 | -0.6734 | Yes | ||
| 52 | FRMD4A | 2190176 | 16646 | -0.255 | -0.6666 | Yes | ||
| 53 | ETV6 | 610524 | 16718 | -0.268 | -0.6594 | Yes | ||
| 54 | KIT | 7040095 | 16733 | -0.270 | -0.6491 | Yes | ||
| 55 | SMAD2 | 4200592 | 16785 | -0.281 | -0.6402 | Yes | ||
| 56 | PERQ1 | 1340164 | 16937 | -0.320 | -0.6352 | Yes | ||
| 57 | NR1H2 | 4070056 | 17065 | -0.359 | -0.6273 | Yes | ||
| 58 | JARID1B | 4730494 | 17254 | -0.435 | -0.6195 | Yes | ||
| 59 | XYLT2 | 6380039 | 17310 | -0.459 | -0.6036 | Yes | ||
| 60 | FCHSD2 | 5720092 | 17525 | -0.558 | -0.5922 | Yes | ||
| 61 | WSB1 | 870563 | 17558 | -0.571 | -0.5704 | Yes | ||
| 62 | ARHGEF5 | 3170605 | 17597 | -0.587 | -0.5483 | Yes | ||
| 63 | PRKACB | 4210170 | 17608 | -0.594 | -0.5244 | Yes | ||
| 64 | RAB5C | 1990397 | 17669 | -0.628 | -0.5017 | Yes | ||
| 65 | GAB1 | 2970156 | 17684 | -0.634 | -0.4764 | Yes | ||
| 66 | CLK2 | 510079 2630736 | 17817 | -0.730 | -0.4534 | Yes | ||
| 67 | NCOA1 | 3610438 | 17841 | -0.745 | -0.4240 | Yes | ||
| 68 | RUNX1 | 3840711 | 17925 | -0.807 | -0.3952 | Yes | ||
| 69 | IRF2 | 5390044 | 18019 | -0.899 | -0.3633 | Yes | ||
| 70 | TRIB2 | 4120605 | 18126 | -1.035 | -0.3263 | Yes | ||
| 71 | PACSIN1 | 3870632 6370021 6900707 | 18140 | -1.047 | -0.2839 | Yes | ||
| 72 | ANKRD50 | 870543 | 18166 | -1.089 | -0.2404 | Yes | ||
| 73 | STK4 | 2640152 | 18203 | -1.158 | -0.1947 | Yes | ||
| 74 | NEDD9 | 1740373 4230053 | 18330 | -1.454 | -0.1416 | Yes | ||
| 75 | PHF2 | 4060300 | 18416 | -1.760 | -0.0738 | Yes | ||
| 76 | ZFP36L1 | 2510138 4120048 | 18471 | -2.052 | 0.0078 | Yes |

