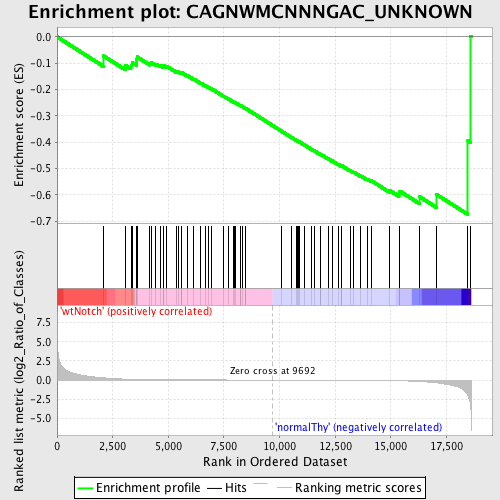
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CAGNWMCNNNGAC_UNKNOWN |
| Enrichment Score (ES) | -0.67499703 |
| Normalized Enrichment Score (NES) | -1.5192358 |
| Nominal p-value | 0.008264462 |
| FDR q-value | 0.1258335 |
| FWER p-Value | 0.716 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NEFH | 630239 | 2086 | 0.286 | -0.0725 | No | ||
| 2 | FGD2 | 5340195 | 3072 | 0.123 | -0.1084 | No | ||
| 3 | RPL4 | 2940014 5420324 | 3322 | 0.099 | -0.1079 | No | ||
| 4 | PTPRN | 5900577 | 3372 | 0.094 | -0.0974 | No | ||
| 5 | DNER | 3610170 | 3576 | 0.079 | -0.0973 | No | ||
| 6 | SLC25A28 | 6590600 | 3580 | 0.079 | -0.0864 | No | ||
| 7 | TNFRSF19 | 3870731 6940730 | 3591 | 0.078 | -0.0760 | No | ||
| 8 | CHGB | 1980053 | 4167 | 0.047 | -0.1004 | No | ||
| 9 | AOC2 | 520348 3120008 | 4230 | 0.045 | -0.0975 | No | ||
| 10 | CHKA | 510324 | 4442 | 0.039 | -0.1034 | No | ||
| 11 | GRM1 | 5890324 | 4628 | 0.035 | -0.1085 | No | ||
| 12 | CACNA1B | 1740010 5360593 | 4773 | 0.032 | -0.1119 | No | ||
| 13 | CHRNB2 | 580204 3120739 | 4792 | 0.031 | -0.1085 | No | ||
| 14 | PAQR4 | 730736 | 4916 | 0.029 | -0.1111 | No | ||
| 15 | LHX3 | 670164 1770280 | 5383 | 0.022 | -0.1331 | No | ||
| 16 | MAPK11 | 130452 3120440 3610465 | 5433 | 0.022 | -0.1327 | No | ||
| 17 | MGAT5B | 380131 2570364 | 5588 | 0.020 | -0.1382 | No | ||
| 18 | SYT6 | 2510280 3850128 4540064 | 5610 | 0.020 | -0.1365 | No | ||
| 19 | PLAC1 | 2350594 | 5847 | 0.017 | -0.1468 | No | ||
| 20 | CHAT | 6840603 | 6142 | 0.015 | -0.1605 | No | ||
| 21 | CALB1 | 460070 | 6465 | 0.013 | -0.1761 | No | ||
| 22 | NPPB | 4150722 | 6684 | 0.011 | -0.1863 | No | ||
| 23 | PTHR1 | 2640711 3360647 | 6793 | 0.011 | -0.1906 | No | ||
| 24 | NHLH2 | 6200021 | 6946 | 0.010 | -0.1974 | No | ||
| 25 | VIP | 2850647 | 7484 | 0.008 | -0.2253 | No | ||
| 26 | SLC12A5 | 1980692 | 7708 | 0.007 | -0.2363 | No | ||
| 27 | RPH3A | 2190156 | 7949 | 0.006 | -0.2485 | No | ||
| 28 | OMG | 6760066 | 7972 | 0.006 | -0.2488 | No | ||
| 29 | BDNF | 2940128 3520368 | 8004 | 0.006 | -0.2497 | No | ||
| 30 | SCHIP1 | 4560152 | 8240 | 0.005 | -0.2617 | No | ||
| 31 | TNRC4 | 4050156 | 8253 | 0.005 | -0.2617 | No | ||
| 32 | POU4F3 | 2690035 | 8313 | 0.004 | -0.2643 | No | ||
| 33 | CHGA | 1990056 6550463 | 8462 | 0.004 | -0.2717 | No | ||
| 34 | SLC17A6 | 4210576 | 8483 | 0.004 | -0.2723 | No | ||
| 35 | GRIN1 | 3800014 7000609 | 10091 | -0.001 | -0.3587 | No | ||
| 36 | CRH | 3710301 | 10535 | -0.003 | -0.3822 | No | ||
| 37 | PAFAH1B1 | 4230333 6420121 6450066 | 10752 | -0.003 | -0.3934 | No | ||
| 38 | PGF | 4810593 | 10786 | -0.003 | -0.3947 | No | ||
| 39 | KCNIP2 | 60088 1780324 | 10844 | -0.004 | -0.3973 | No | ||
| 40 | GLRA3 | 2570500 | 10902 | -0.004 | -0.3999 | No | ||
| 41 | BARHL1 | 4210113 | 11107 | -0.004 | -0.4102 | No | ||
| 42 | HTR1A | 6980441 | 11449 | -0.006 | -0.4278 | No | ||
| 43 | DRD3 | 4780402 | 11570 | -0.006 | -0.4335 | No | ||
| 44 | COL5A3 | 1940471 | 11818 | -0.007 | -0.4458 | No | ||
| 45 | HPCA | 6860010 | 11854 | -0.007 | -0.4467 | No | ||
| 46 | DBC1 | 4610156 | 12181 | -0.009 | -0.4630 | No | ||
| 47 | JMJD1C | 940575 2120025 | 12357 | -0.010 | -0.4711 | No | ||
| 48 | MYF5 | 1050739 | 12653 | -0.011 | -0.4854 | No | ||
| 49 | ELAVL3 | 2850014 | 12764 | -0.012 | -0.4897 | No | ||
| 50 | L1CAM | 4850021 | 12783 | -0.012 | -0.4890 | No | ||
| 51 | GABRG2 | 2350402 4210204 6130279 6550037 | 13172 | -0.015 | -0.5078 | No | ||
| 52 | DRD2 | 5890369 | 13329 | -0.016 | -0.5139 | No | ||
| 53 | PHYHIPL | 2360706 3840692 | 13626 | -0.020 | -0.5271 | No | ||
| 54 | SYT4 | 5080193 | 13935 | -0.024 | -0.5404 | No | ||
| 55 | SCN3B | 840112 1780152 5910685 6100131 | 14120 | -0.027 | -0.5465 | No | ||
| 56 | CSMD3 | 1660427 1940687 | 14940 | -0.052 | -0.5833 | No | ||
| 57 | PRG3 | 6220020 | 15368 | -0.078 | -0.5955 | No | ||
| 58 | TCF1 | 5390022 | 15393 | -0.079 | -0.5858 | No | ||
| 59 | WAS | 5270193 | 16305 | -0.192 | -0.6081 | Yes | ||
| 60 | PFN1 | 6130132 | 17047 | -0.354 | -0.5986 | Yes | ||
| 61 | CDK5R1 | 3870161 | 18465 | -2.008 | -0.3949 | Yes | ||
| 62 | MYLC2PL | 360280 | 18564 | -2.889 | 0.0028 | Yes |

