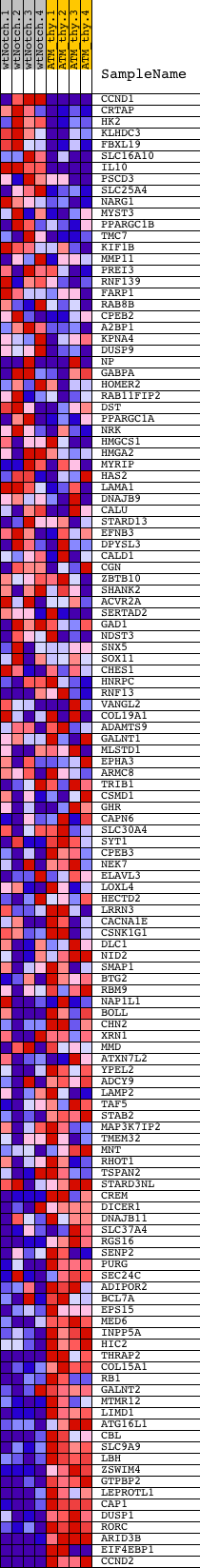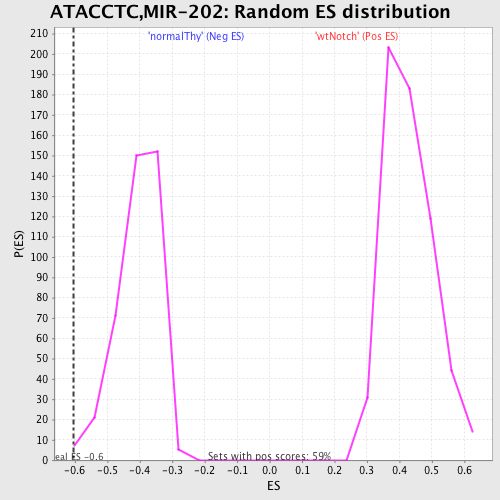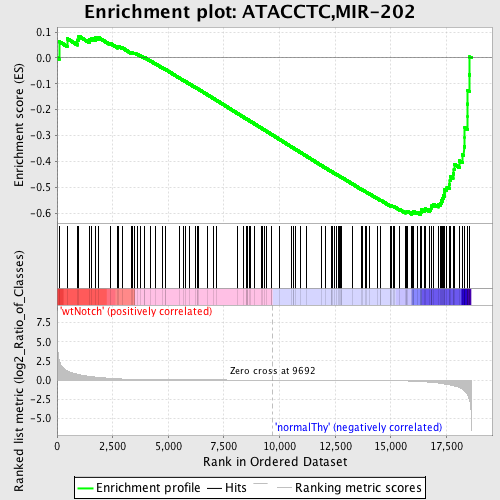
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | ATACCTC,MIR-202 |
| Enrichment Score (ES) | -0.6053372 |
| Normalized Enrichment Score (NES) | -1.4970772 |
| Nominal p-value | 0.0049261083 |
| FDR q-value | 0.15524596 |
| FWER p-Value | 0.831 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCND1 | 460524 770309 3120576 6980398 | 90 | 2.631 | 0.0633 | No | ||
| 2 | CRTAP | 3990725 | 452 | 1.189 | 0.0746 | No | ||
| 3 | HK2 | 2640722 | 915 | 0.752 | 0.0691 | No | ||
| 4 | KLHDC3 | 3830717 | 975 | 0.700 | 0.0841 | No | ||
| 5 | FBXL19 | 4570039 | 1453 | 0.478 | 0.0707 | No | ||
| 6 | SLC16A10 | 4010672 | 1553 | 0.437 | 0.0767 | No | ||
| 7 | IL10 | 2340685 2640541 2850403 6590286 | 1724 | 0.384 | 0.0774 | No | ||
| 8 | PSCD3 | 730301 | 1869 | 0.343 | 0.0785 | No | ||
| 9 | SLC25A4 | 2360519 | 2399 | 0.217 | 0.0555 | No | ||
| 10 | NARG1 | 5910563 6350095 | 2725 | 0.163 | 0.0422 | No | ||
| 11 | MYST3 | 5270500 | 2753 | 0.158 | 0.0448 | No | ||
| 12 | PPARGC1B | 3290114 | 2930 | 0.136 | 0.0388 | No | ||
| 13 | TMC7 | 1990129 5290253 | 3326 | 0.099 | 0.0200 | No | ||
| 14 | KIF1B | 1240494 2370139 4570270 6510102 | 3385 | 0.093 | 0.0193 | No | ||
| 15 | MMP11 | 1980619 | 3465 | 0.088 | 0.0173 | No | ||
| 16 | PREI3 | 3170066 3440075 6370427 | 3481 | 0.087 | 0.0187 | No | ||
| 17 | RNF139 | 2650114 3610450 | 3593 | 0.078 | 0.0148 | No | ||
| 18 | FARP1 | 5290102 7100341 | 3744 | 0.067 | 0.0084 | No | ||
| 19 | RAB8B | 510692 1980017 3130136 4570739 | 3930 | 0.058 | -0.0001 | No | ||
| 20 | CPEB2 | 4760338 | 3936 | 0.057 | 0.0011 | No | ||
| 21 | A2BP1 | 2370390 4590593 5550014 | 4218 | 0.046 | -0.0129 | No | ||
| 22 | KPNA4 | 510369 | 4400 | 0.040 | -0.0217 | No | ||
| 23 | DUSP9 | 630605 | 4722 | 0.033 | -0.0382 | No | ||
| 24 | NP | 2630577 3120142 5670068 6290039 | 4866 | 0.030 | -0.0451 | No | ||
| 25 | GABPA | 2360017 4780301 4850279 | 4872 | 0.030 | -0.0446 | No | ||
| 26 | HOMER2 | 840242 | 5501 | 0.021 | -0.0780 | No | ||
| 27 | RAB11FIP2 | 4570040 | 5670 | 0.019 | -0.0866 | No | ||
| 28 | DST | 430026 1090035 2340577 3170068 3870112 4780519 6400167 6450358 7040347 | 5780 | 0.018 | -0.0921 | No | ||
| 29 | PPARGC1A | 4670040 | 5938 | 0.017 | -0.1001 | No | ||
| 30 | NRK | 4590609 | 6229 | 0.014 | -0.1154 | No | ||
| 31 | HMGCS1 | 6620452 | 6288 | 0.014 | -0.1182 | No | ||
| 32 | HMGA2 | 2940121 3390647 5130279 6400136 | 6327 | 0.014 | -0.1199 | No | ||
| 33 | MYRIP | 1580471 | 6350 | 0.014 | -0.1207 | No | ||
| 34 | HAS2 | 5360181 | 6764 | 0.011 | -0.1428 | No | ||
| 35 | LAMA1 | 1770594 | 7025 | 0.010 | -0.1566 | No | ||
| 36 | DNAJB9 | 460114 | 7148 | 0.009 | -0.1630 | No | ||
| 37 | CALU | 2680403 3520056 4610348 5720176 6130121 | 8093 | 0.005 | -0.2139 | No | ||
| 38 | STARD13 | 540722 | 8398 | 0.004 | -0.2302 | No | ||
| 39 | EFNB3 | 5570594 | 8508 | 0.004 | -0.2360 | No | ||
| 40 | DPYSL3 | 1580594 | 8523 | 0.004 | -0.2367 | No | ||
| 41 | CALD1 | 1770129 1940397 | 8553 | 0.004 | -0.2382 | No | ||
| 42 | CGN | 5890139 6370167 6400537 | 8556 | 0.004 | -0.2382 | No | ||
| 43 | ZBTB10 | 4200132 | 8667 | 0.003 | -0.2440 | No | ||
| 44 | SHANK2 | 1500452 | 8693 | 0.003 | -0.2453 | No | ||
| 45 | ACVR2A | 6110647 | 8884 | 0.002 | -0.2555 | No | ||
| 46 | SERTAD2 | 2810673 6840450 | 9202 | 0.001 | -0.2726 | No | ||
| 47 | GAD1 | 2360035 3140167 | 9223 | 0.001 | -0.2737 | No | ||
| 48 | NDST3 | 670020 | 9321 | 0.001 | -0.2789 | No | ||
| 49 | SNX5 | 1570021 6860609 | 9427 | 0.001 | -0.2846 | No | ||
| 50 | SOX11 | 610279 | 9618 | 0.000 | -0.2948 | No | ||
| 51 | CHES1 | 4570121 5700162 | 9641 | 0.000 | -0.2960 | No | ||
| 52 | HNRPC | 1570133 3610131 | 9993 | -0.001 | -0.3150 | No | ||
| 53 | RNF13 | 2370021 | 10539 | -0.003 | -0.3444 | No | ||
| 54 | VANGL2 | 70097 870075 | 10606 | -0.003 | -0.3479 | No | ||
| 55 | COL19A1 | 6220441 | 10646 | -0.003 | -0.3499 | No | ||
| 56 | ADAMTS9 | 2760079 | 10728 | -0.003 | -0.3542 | No | ||
| 57 | GALNT1 | 4070487 6130100 6250132 6900048 | 10920 | -0.004 | -0.3645 | No | ||
| 58 | MLSTD1 | 3710184 | 11209 | -0.005 | -0.3799 | No | ||
| 59 | EPHA3 | 2190497 5080059 | 11898 | -0.007 | -0.4169 | No | ||
| 60 | ARMC8 | 520408 940132 4060368 4210102 | 12056 | -0.008 | -0.4252 | No | ||
| 61 | TRIB1 | 2320435 | 12344 | -0.010 | -0.4405 | No | ||
| 62 | CSMD1 | 1780672 | 12352 | -0.010 | -0.4406 | No | ||
| 63 | GHR | 2260156 | 12358 | -0.010 | -0.4406 | No | ||
| 64 | CAPN6 | 1740168 | 12464 | -0.010 | -0.4461 | No | ||
| 65 | SLC30A4 | 1850372 | 12571 | -0.011 | -0.4515 | No | ||
| 66 | SYT1 | 840364 | 12633 | -0.011 | -0.4545 | No | ||
| 67 | CPEB3 | 3940164 | 12700 | -0.011 | -0.4578 | No | ||
| 68 | NEK7 | 4540026 | 12718 | -0.012 | -0.4584 | No | ||
| 69 | ELAVL3 | 2850014 | 12764 | -0.012 | -0.4605 | No | ||
| 70 | LOXL4 | 6520095 | 12795 | -0.012 | -0.4619 | No | ||
| 71 | HECTD2 | 2680014 3060689 | 13293 | -0.016 | -0.4883 | No | ||
| 72 | LRRN3 | 1450133 | 13682 | -0.020 | -0.5088 | No | ||
| 73 | CACNA1E | 840253 | 13704 | -0.021 | -0.5094 | No | ||
| 74 | CSNK1G1 | 840082 1230575 3940647 | 13858 | -0.023 | -0.5171 | No | ||
| 75 | DLC1 | 1090632 6450594 | 13911 | -0.024 | -0.5193 | No | ||
| 76 | NID2 | 2320059 | 14023 | -0.026 | -0.5246 | No | ||
| 77 | SMAP1 | 840056 2360523 | 14384 | -0.033 | -0.5432 | No | ||
| 78 | BTG2 | 2350411 | 14531 | -0.036 | -0.5502 | No | ||
| 79 | RBM9 | 2510092 6290088 | 14552 | -0.037 | -0.5503 | No | ||
| 80 | NAP1L1 | 2510593 5900215 5090470 450079 4920280 | 14974 | -0.054 | -0.5717 | No | ||
| 81 | BOLL | 6100743 | 14997 | -0.054 | -0.5715 | No | ||
| 82 | CHN2 | 870528 | 15031 | -0.056 | -0.5718 | No | ||
| 83 | XRN1 | 5900093 | 15097 | -0.060 | -0.5738 | No | ||
| 84 | MMD | 3850239 | 15143 | -0.063 | -0.5746 | No | ||
| 85 | ATXN7L2 | 4010161 | 15407 | -0.081 | -0.5867 | No | ||
| 86 | YPEL2 | 5560609 | 15649 | -0.105 | -0.5970 | No | ||
| 87 | ADCY9 | 5690168 | 15689 | -0.108 | -0.5963 | No | ||
| 88 | LAMP2 | 1230402 1980373 | 15722 | -0.111 | -0.5952 | No | ||
| 89 | TAF5 | 3450288 5890193 6860435 | 15767 | -0.117 | -0.5945 | No | ||
| 90 | STAB2 | 4810452 | 15944 | -0.140 | -0.6004 | Yes | ||
| 91 | MAP3K7IP2 | 2340242 | 15958 | -0.142 | -0.5974 | Yes | ||
| 92 | TMEM32 | 1170452 | 16008 | -0.149 | -0.5962 | Yes | ||
| 93 | MNT | 4920575 | 16026 | -0.150 | -0.5932 | Yes | ||
| 94 | RHOT1 | 6760692 | 16211 | -0.178 | -0.5986 | Yes | ||
| 95 | TSPAN2 | 3940161 | 16337 | -0.198 | -0.6002 | Yes | ||
| 96 | STARD3NL | 7000114 | 16354 | -0.202 | -0.5958 | Yes | ||
| 97 | CREM | 840156 6380438 6660041 6660168 | 16364 | -0.204 | -0.5910 | Yes | ||
| 98 | DICER1 | 540286 | 16382 | -0.206 | -0.5866 | Yes | ||
| 99 | DNAJB11 | 4150168 | 16522 | -0.232 | -0.5881 | Yes | ||
| 100 | SLC37A4 | 6400079 | 16551 | -0.238 | -0.5835 | Yes | ||
| 101 | RGS16 | 780091 | 16741 | -0.273 | -0.5866 | Yes | ||
| 102 | SENP2 | 4540452 | 16805 | -0.287 | -0.5826 | Yes | ||
| 103 | PURG | 1190593 | 16813 | -0.290 | -0.5755 | Yes | ||
| 104 | SEC24C | 6980707 | 16844 | -0.299 | -0.5693 | Yes | ||
| 105 | ADIPOR2 | 1510035 | 16917 | -0.317 | -0.5650 | Yes | ||
| 106 | BCL7A | 4560095 | 17129 | -0.383 | -0.5665 | Yes | ||
| 107 | EPS15 | 4200215 | 17238 | -0.428 | -0.5613 | Yes | ||
| 108 | MED6 | 6840500 | 17267 | -0.439 | -0.5514 | Yes | ||
| 109 | INPP5A | 5910195 | 17315 | -0.462 | -0.5420 | Yes | ||
| 110 | HIC2 | 4010433 | 17372 | -0.492 | -0.5322 | Yes | ||
| 111 | THRAP2 | 4920600 | 17418 | -0.510 | -0.5215 | Yes | ||
| 112 | COL15A1 | 6840113 | 17424 | -0.512 | -0.5085 | Yes | ||
| 113 | RB1 | 5900338 | 17522 | -0.556 | -0.4993 | Yes | ||
| 114 | GALNT2 | 2260279 4060239 | 17642 | -0.612 | -0.4899 | Yes | ||
| 115 | MTMR12 | 7000181 | 17658 | -0.622 | -0.4745 | Yes | ||
| 116 | LIMD1 | 2480142 | 17680 | -0.632 | -0.4593 | Yes | ||
| 117 | ATG16L1 | 5390292 | 17796 | -0.714 | -0.4470 | Yes | ||
| 118 | CBL | 6380068 | 17838 | -0.744 | -0.4299 | Yes | ||
| 119 | SLC9A9 | 1940309 | 17850 | -0.753 | -0.4110 | Yes | ||
| 120 | LBH | 70093 | 18081 | -0.978 | -0.3981 | Yes | ||
| 121 | ZSWIM4 | 6940195 | 18225 | -1.202 | -0.3747 | Yes | ||
| 122 | GTPBP2 | 2760750 | 18289 | -1.353 | -0.3430 | Yes | ||
| 123 | LEPROTL1 | 6900110 | 18323 | -1.434 | -0.3076 | Yes | ||
| 124 | CAP1 | 2650278 | 18327 | -1.446 | -0.2703 | Yes | ||
| 125 | DUSP1 | 6860121 | 18444 | -1.881 | -0.2278 | Yes | ||
| 126 | RORC | 1740121 | 18449 | -1.922 | -0.1782 | Yes | ||
| 127 | ARID3B | 6650008 | 18459 | -1.981 | -0.1273 | Yes | ||
| 128 | EIF4EBP1 | 60132 5720148 | 18525 | -2.523 | -0.0655 | Yes | ||
| 129 | CCND2 | 5340167 | 18548 | -2.713 | 0.0037 | Yes |