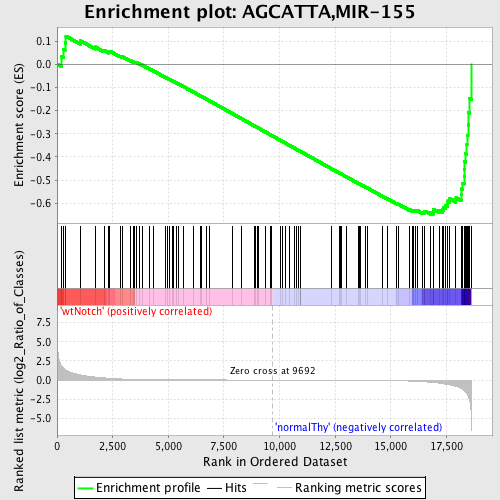
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | AGCATTA,MIR-155 |
| Enrichment Score (ES) | -0.64891106 |
| Normalized Enrichment Score (NES) | -1.56368 |
| Nominal p-value | 0.0025252525 |
| FDR q-value | 0.15563498 |
| FWER p-Value | 0.478 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSIP1 | 1780082 3190435 5050594 | 201 | 1.844 | 0.0322 | No | ||
| 2 | CEBPB | 2970019 | 282 | 1.602 | 0.0652 | No | ||
| 3 | BCAP29 | 2350500 | 357 | 1.383 | 0.0935 | No | ||
| 4 | BCAT1 | 3290128 4050408 | 397 | 1.300 | 0.1217 | No | ||
| 5 | TIMM8A | 110279 | 1057 | 0.652 | 0.1013 | No | ||
| 6 | SPI1 | 1410397 | 1713 | 0.386 | 0.0749 | No | ||
| 7 | RPS6KA3 | 1980707 | 2107 | 0.281 | 0.0603 | No | ||
| 8 | UBE2D3 | 3190452 | 2324 | 0.231 | 0.0540 | No | ||
| 9 | CUGBP2 | 6180121 6840139 | 2374 | 0.221 | 0.0565 | No | ||
| 10 | YWHAZ | 1230717 | 2828 | 0.149 | 0.0355 | No | ||
| 11 | CSF1R | 2340110 6420408 | 2951 | 0.134 | 0.0320 | No | ||
| 12 | SLC39A10 | 4150347 | 3307 | 0.101 | 0.0152 | No | ||
| 13 | NFAT5 | 2510411 5890195 6550152 | 3440 | 0.090 | 0.0102 | No | ||
| 14 | STRN3 | 1450093 6200685 | 3486 | 0.086 | 0.0098 | No | ||
| 15 | KRAS | 2060170 | 3552 | 0.081 | 0.0082 | No | ||
| 16 | PKIA | 4070441 | 3684 | 0.072 | 0.0027 | No | ||
| 17 | ETS1 | 5270278 6450717 6620465 | 3819 | 0.063 | -0.0030 | No | ||
| 18 | CDC73 | 1580132 | 4168 | 0.047 | -0.0207 | No | ||
| 19 | SCG2 | 5670438 130671 | 4341 | 0.042 | -0.0290 | No | ||
| 20 | UPP2 | 2190091 2680746 6940647 | 4886 | 0.030 | -0.0577 | No | ||
| 21 | SEMA5A | 6200110 6520435 | 4968 | 0.028 | -0.0614 | No | ||
| 22 | QKI | 5220093 6130044 | 5061 | 0.027 | -0.0658 | No | ||
| 23 | DYNC1I1 | 1410605 | 5168 | 0.025 | -0.0709 | No | ||
| 24 | RAB1A | 2370671 | 5240 | 0.024 | -0.0742 | No | ||
| 25 | CNTN4 | 1780300 5570577 6370019 | 5387 | 0.022 | -0.0815 | No | ||
| 26 | LRP1B | 1450193 | 5456 | 0.022 | -0.0847 | No | ||
| 27 | RAB11FIP2 | 4570040 | 5670 | 0.019 | -0.0958 | No | ||
| 28 | HIVEP2 | 6520538 | 6148 | 0.015 | -0.1212 | No | ||
| 29 | NDFIP1 | 1980402 | 6449 | 0.013 | -0.1371 | No | ||
| 30 | PICALM | 1940280 | 6478 | 0.013 | -0.1383 | No | ||
| 31 | SGCZ | 3840592 | 6715 | 0.011 | -0.1508 | No | ||
| 32 | SERAC1 | 2630220 | 6854 | 0.010 | -0.1580 | No | ||
| 33 | CAB39 | 5720471 | 7869 | 0.006 | -0.2126 | No | ||
| 34 | MAP3K10 | 3360168 4850180 | 8277 | 0.005 | -0.2345 | No | ||
| 35 | ACVR2A | 6110647 | 8884 | 0.002 | -0.2671 | No | ||
| 36 | RCN2 | 840324 | 8900 | 0.002 | -0.2679 | No | ||
| 37 | TOMM20 | 6510113 | 9011 | 0.002 | -0.2738 | No | ||
| 38 | PTPN2 | 130315 | 9056 | 0.002 | -0.2761 | No | ||
| 39 | FGF7 | 5390484 | 9370 | 0.001 | -0.2930 | No | ||
| 40 | GDF6 | 1340048 | 9571 | 0.000 | -0.3038 | No | ||
| 41 | SOX11 | 610279 | 9618 | 0.000 | -0.3063 | No | ||
| 42 | TRIM2 | 430546 1170286 | 10027 | -0.001 | -0.3283 | No | ||
| 43 | YWHAE | 5310435 | 10135 | -0.001 | -0.3340 | No | ||
| 44 | BOC | 5270348 | 10255 | -0.002 | -0.3404 | No | ||
| 45 | HIF1A | 5670605 | 10440 | -0.002 | -0.3503 | No | ||
| 46 | EDG1 | 4200619 | 10690 | -0.003 | -0.3637 | No | ||
| 47 | ZIC3 | 380020 | 10740 | -0.003 | -0.3662 | No | ||
| 48 | ELL2 | 6420091 | 10843 | -0.004 | -0.3717 | No | ||
| 49 | SP3 | 3840338 | 10919 | -0.004 | -0.3756 | No | ||
| 50 | GPM6B | 6220458 | 10936 | -0.004 | -0.3764 | No | ||
| 51 | ARID2 | 110647 5390435 | 10953 | -0.004 | -0.3772 | No | ||
| 52 | SOX1 | 630026 | 12353 | -0.010 | -0.4525 | No | ||
| 53 | MARCH7 | 3800020 6620168 | 12703 | -0.011 | -0.4711 | No | ||
| 54 | OGN | 3710326 3800193 | 12733 | -0.012 | -0.4724 | No | ||
| 55 | MYO1D | 6760292 | 12767 | -0.012 | -0.4739 | No | ||
| 56 | WEE1 | 3390070 | 13018 | -0.014 | -0.4870 | No | ||
| 57 | DNAJB7 | 1090593 | 13536 | -0.018 | -0.5145 | No | ||
| 58 | MEIS1 | 1400575 | 13595 | -0.019 | -0.5172 | No | ||
| 59 | PCDH9 | 2100136 | 13631 | -0.020 | -0.5187 | No | ||
| 60 | ACTA1 | 840538 | 13860 | -0.023 | -0.5304 | No | ||
| 61 | BCORL1 | 1660008 | 13941 | -0.024 | -0.5342 | No | ||
| 62 | ARL5B | 3840059 | 14627 | -0.039 | -0.5703 | No | ||
| 63 | GPR85 | 5130750 5910020 | 14835 | -0.047 | -0.5803 | No | ||
| 64 | RAB6A | 3840463 4780731 | 15250 | -0.070 | -0.6011 | No | ||
| 65 | MYO10 | 3830576 5570092 | 15344 | -0.076 | -0.6043 | No | ||
| 66 | FBXO33 | 1510504 | 15847 | -0.127 | -0.6285 | No | ||
| 67 | MAP3K7IP2 | 2340242 | 15958 | -0.142 | -0.6311 | No | ||
| 68 | CSNK1G2 | 3060095 4730037 | 16010 | -0.149 | -0.6304 | No | ||
| 69 | SGK3 | 1990408 4760452 5130278 6020594 6620471 | 16103 | -0.163 | -0.6315 | No | ||
| 70 | NFIX | 2450152 | 16214 | -0.179 | -0.6333 | No | ||
| 71 | SYPL1 | 2680706 5900403 | 16433 | -0.217 | -0.6400 | No | ||
| 72 | SKI | 6590008 | 16516 | -0.231 | -0.6391 | No | ||
| 73 | MECP2 | 1940450 | 16526 | -0.233 | -0.6341 | No | ||
| 74 | CHD7 | 3870372 | 16801 | -0.286 | -0.6422 | Yes | ||
| 75 | KCNN3 | 6520600 6420138 | 16898 | -0.313 | -0.6401 | Yes | ||
| 76 | SSH2 | 360056 | 16905 | -0.314 | -0.6331 | Yes | ||
| 77 | H3F3A | 2900086 | 16916 | -0.317 | -0.6263 | Yes | ||
| 78 | BRD1 | 1660278 | 17177 | -0.402 | -0.6310 | Yes | ||
| 79 | IKBKE | 1850332 | 17319 | -0.463 | -0.6278 | Yes | ||
| 80 | MAP3K14 | 5890435 | 17378 | -0.493 | -0.6194 | Yes | ||
| 81 | MYB | 1660494 5860451 6130706 | 17459 | -0.526 | -0.6114 | Yes | ||
| 82 | SMARCA4 | 6980528 | 17543 | -0.565 | -0.6027 | Yes | ||
| 83 | SLA | 2000070 | 17568 | -0.575 | -0.5906 | Yes | ||
| 84 | BACH1 | 290195 | 17652 | -0.618 | -0.5807 | Yes | ||
| 85 | CARHSP1 | 3390402 | 17927 | -0.808 | -0.5766 | Yes | ||
| 86 | DNAJB1 | 1090041 | 18169 | -1.092 | -0.5642 | Yes | ||
| 87 | SOCS1 | 730139 | 18178 | -1.112 | -0.5387 | Yes | ||
| 88 | CD47 | 1850603 | 18214 | -1.182 | -0.5130 | Yes | ||
| 89 | KBTBD2 | 2320014 | 18294 | -1.366 | -0.4854 | Yes | ||
| 90 | INPP5D | 3450731 | 18308 | -1.388 | -0.4537 | Yes | ||
| 91 | SATB1 | 5670154 | 18320 | -1.430 | -0.4210 | Yes | ||
| 92 | CTLA4 | 6590537 | 18369 | -1.563 | -0.3871 | Yes | ||
| 93 | SDCBP | 460487 2190039 5270441 | 18420 | -1.784 | -0.3482 | Yes | ||
| 94 | HBP1 | 2510102 3130010 4210619 | 18428 | -1.800 | -0.3066 | Yes | ||
| 95 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 18469 | -2.031 | -0.2614 | Yes | ||
| 96 | ITK | 2230592 | 18500 | -2.294 | -0.2095 | Yes | ||
| 97 | TLE4 | 1450064 1500519 | 18549 | -2.720 | -0.1486 | Yes | ||
| 98 | MIDN | 2260132 5220377 | 18616 | -6.526 | -0.0000 | Yes |

