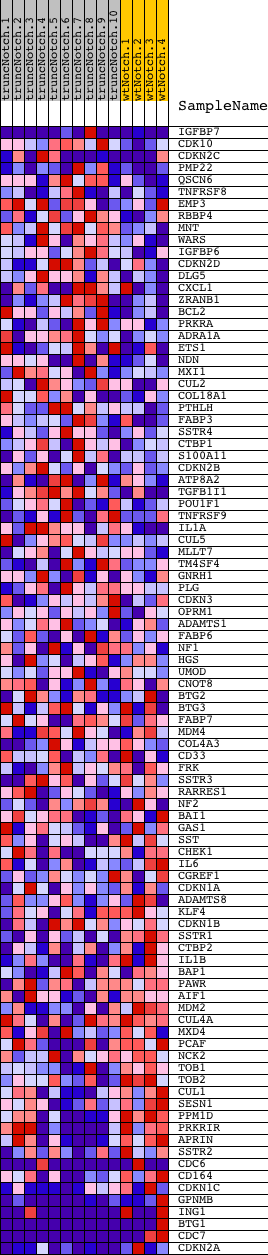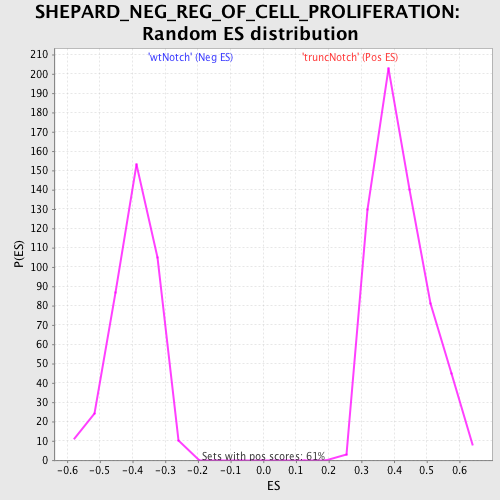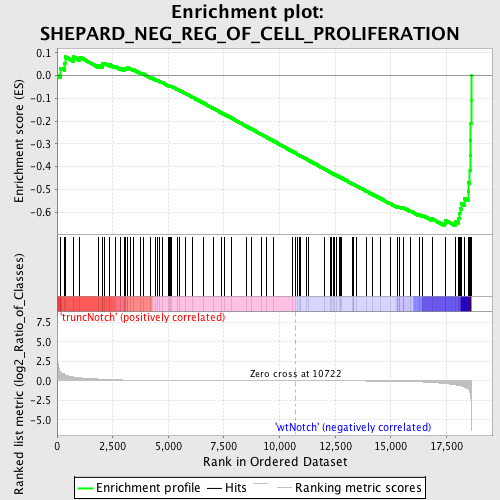
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | SHEPARD_NEG_REG_OF_CELL_PROLIFERATION |
| Enrichment Score (ES) | -0.6607956 |
| Normalized Enrichment Score (NES) | -1.6759448 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1289581 |
| FWER p-Value | 0.608 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IGFBP7 | 520411 3060110 5290152 | 166 | 1.108 | 0.0331 | No | ||
| 2 | CDK10 | 60164 1190717 3130128 3990044 | 344 | 0.815 | 0.0545 | No | ||
| 3 | CDKN2C | 5050750 5130148 | 376 | 0.763 | 0.0818 | No | ||
| 4 | PMP22 | 4010239 6550072 | 721 | 0.499 | 0.0821 | No | ||
| 5 | QSCN6 | 3120441 | 1026 | 0.393 | 0.0806 | No | ||
| 6 | TNFRSF8 | 870136 4810446 | 1850 | 0.237 | 0.0452 | No | ||
| 7 | EMP3 | 1500722 1570039 | 2042 | 0.210 | 0.0429 | No | ||
| 8 | RBBP4 | 6650528 | 2043 | 0.210 | 0.0509 | No | ||
| 9 | MNT | 4920575 | 2116 | 0.200 | 0.0546 | No | ||
| 10 | WARS | 2570717 3290139 | 2368 | 0.168 | 0.0474 | No | ||
| 11 | IGFBP6 | 1580181 | 2623 | 0.140 | 0.0390 | No | ||
| 12 | CDKN2D | 6040035 | 2854 | 0.118 | 0.0310 | No | ||
| 13 | DLG5 | 450215 | 3034 | 0.104 | 0.0253 | No | ||
| 14 | CXCL1 | 2690537 | 3037 | 0.104 | 0.0292 | No | ||
| 15 | ZRANB1 | 510373 4540278 | 3083 | 0.102 | 0.0306 | No | ||
| 16 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 3151 | 0.097 | 0.0307 | No | ||
| 17 | PRKRA | 2510600 | 3154 | 0.097 | 0.0343 | No | ||
| 18 | ADRA1A | 1230446 2260390 | 3302 | 0.088 | 0.0297 | No | ||
| 19 | ETS1 | 5270278 6450717 6620465 | 3446 | 0.079 | 0.0250 | No | ||
| 20 | NDN | 5670075 | 3761 | 0.066 | 0.0105 | No | ||
| 21 | MXI1 | 5050064 5130484 | 3870 | 0.061 | 0.0070 | No | ||
| 22 | CUL2 | 4200278 | 4216 | 0.051 | -0.0097 | No | ||
| 23 | COL18A1 | 610301 4570338 | 4405 | 0.046 | -0.0181 | No | ||
| 24 | PTHLH | 5290739 | 4492 | 0.044 | -0.0211 | No | ||
| 25 | FABP3 | 6860452 | 4588 | 0.042 | -0.0246 | No | ||
| 26 | SSTR4 | 6100398 | 4736 | 0.039 | -0.0311 | No | ||
| 27 | CTBP1 | 3780315 | 5025 | 0.033 | -0.0454 | No | ||
| 28 | S100A11 | 2260064 | 5045 | 0.033 | -0.0451 | No | ||
| 29 | CDKN2B | 6020040 | 5117 | 0.032 | -0.0478 | No | ||
| 30 | ATP8A2 | 1580086 3940025 | 5131 | 0.032 | -0.0473 | No | ||
| 31 | TGFB1I1 | 2060288 6550450 | 5401 | 0.028 | -0.0608 | No | ||
| 32 | POU1F1 | 450148 | 5519 | 0.026 | -0.0661 | No | ||
| 33 | TNFRSF9 | 2510400 6650484 | 5770 | 0.023 | -0.0787 | No | ||
| 34 | IL1A | 3610056 | 6101 | 0.020 | -0.0957 | No | ||
| 35 | CUL5 | 450142 | 6562 | 0.016 | -0.1200 | No | ||
| 36 | MLLT7 | 4480707 | 7050 | 0.013 | -0.1458 | No | ||
| 37 | TM4SF4 | 540600 | 7389 | 0.011 | -0.1636 | No | ||
| 38 | GNRH1 | 430167 | 7505 | 0.011 | -0.1694 | No | ||
| 39 | PLG | 3360270 3840100 | 7851 | 0.009 | -0.1877 | No | ||
| 40 | CDKN3 | 110520 | 8494 | 0.007 | -0.2221 | No | ||
| 41 | OPRM1 | 5360279 | 8737 | 0.006 | -0.2349 | No | ||
| 42 | ADAMTS1 | 3140286 5860463 | 9197 | 0.004 | -0.2596 | No | ||
| 43 | FABP6 | 7000278 | 9390 | 0.004 | -0.2698 | No | ||
| 44 | NF1 | 6980433 | 9709 | 0.003 | -0.2868 | No | ||
| 45 | HGS | 5570722 | 10569 | 0.000 | -0.3332 | No | ||
| 46 | UMOD | 1980603 | 10730 | -0.000 | -0.3418 | No | ||
| 47 | CNOT8 | 4590129 | 10793 | -0.000 | -0.3452 | No | ||
| 48 | BTG2 | 2350411 | 10910 | -0.001 | -0.3514 | No | ||
| 49 | BTG3 | 7050079 | 10952 | -0.001 | -0.3536 | No | ||
| 50 | FABP7 | 4590142 | 11208 | -0.001 | -0.3673 | No | ||
| 51 | MDM4 | 4070504 4780008 | 11307 | -0.002 | -0.3725 | No | ||
| 52 | COL4A3 | 5910075 | 12009 | -0.004 | -0.4102 | No | ||
| 53 | CD33 | 3990735 | 12271 | -0.005 | -0.4241 | No | ||
| 54 | FRK | 5130333 | 12341 | -0.005 | -0.4277 | No | ||
| 55 | SSTR3 | 5420064 | 12405 | -0.005 | -0.4309 | No | ||
| 56 | RARRES1 | 770139 5860390 | 12469 | -0.006 | -0.4340 | No | ||
| 57 | NF2 | 4150735 6450139 | 12570 | -0.006 | -0.4392 | No | ||
| 58 | BAI1 | 6350053 | 12699 | -0.007 | -0.4459 | No | ||
| 59 | GAS1 | 2120504 | 12734 | -0.007 | -0.4474 | No | ||
| 60 | SST | 6590142 | 12786 | -0.007 | -0.4499 | No | ||
| 61 | CHEK1 | 50167 1940300 | 13273 | -0.010 | -0.4758 | No | ||
| 62 | IL6 | 380133 | 13300 | -0.010 | -0.4768 | No | ||
| 63 | CGREF1 | 4540546 | 13303 | -0.010 | -0.4765 | No | ||
| 64 | CDKN1A | 4050088 6400706 | 13471 | -0.011 | -0.4851 | No | ||
| 65 | ADAMTS8 | 3990497 | 13904 | -0.015 | -0.5078 | No | ||
| 66 | KLF4 | 1850022 3830239 5570750 | 14192 | -0.019 | -0.5226 | No | ||
| 67 | CDKN1B | 3800025 6450044 | 14515 | -0.024 | -0.5391 | No | ||
| 68 | SSTR1 | 2630471 | 14971 | -0.036 | -0.5623 | No | ||
| 69 | CTBP2 | 430309 3710079 | 15290 | -0.049 | -0.5776 | No | ||
| 70 | IL1B | 2640364 | 15295 | -0.049 | -0.5759 | No | ||
| 71 | BAP1 | 3830131 | 15391 | -0.054 | -0.5790 | No | ||
| 72 | PAWR | 5130546 6330706 | 15404 | -0.055 | -0.5776 | No | ||
| 73 | AIF1 | 130332 5220017 | 15550 | -0.064 | -0.5830 | No | ||
| 74 | MDM2 | 3450053 5080138 | 15571 | -0.065 | -0.5816 | No | ||
| 75 | CUL4A | 1170088 2320008 2470278 | 15889 | -0.087 | -0.5954 | No | ||
| 76 | MXD4 | 4810113 | 16283 | -0.123 | -0.6120 | No | ||
| 77 | PCAF | 2230161 2570369 6550451 | 16443 | -0.142 | -0.6152 | No | ||
| 78 | NCK2 | 2510010 | 16852 | -0.206 | -0.6294 | No | ||
| 79 | TOB1 | 4150138 | 17435 | -0.340 | -0.6479 | Yes | ||
| 80 | TOB2 | 1240465 | 17445 | -0.342 | -0.6354 | Yes | ||
| 81 | CUL1 | 1990632 | 17893 | -0.503 | -0.6404 | Yes | ||
| 82 | SESN1 | 2120021 | 18034 | -0.573 | -0.6262 | Yes | ||
| 83 | PPM1D | 1690193 | 18074 | -0.602 | -0.6055 | Yes | ||
| 84 | PRKRIR | 2370402 | 18111 | -0.619 | -0.5839 | Yes | ||
| 85 | APRIN | 2810050 | 18163 | -0.652 | -0.5620 | Yes | ||
| 86 | SSTR2 | 4590687 | 18321 | -0.785 | -0.5406 | Yes | ||
| 87 | CDC6 | 4570296 5360600 | 18471 | -1.030 | -0.5096 | Yes | ||
| 88 | CD164 | 3830594 | 18484 | -1.070 | -0.4696 | Yes | ||
| 89 | CDKN1C | 6520577 | 18557 | -1.527 | -0.4155 | Yes | ||
| 90 | GPNMB | 1780138 2340176 3390403 4560156 | 18576 | -1.698 | -0.3521 | Yes | ||
| 91 | ING1 | 5690010 6520056 | 18583 | -1.765 | -0.2854 | Yes | ||
| 92 | BTG1 | 4200735 6040131 6200133 | 18596 | -1.988 | -0.2106 | Yes | ||
| 93 | CDC7 | 4060546 4850041 | 18609 | -2.689 | -0.1091 | Yes | ||
| 94 | CDKN2A | 4670215 4760047 | 18612 | -2.884 | 0.0002 | Yes |