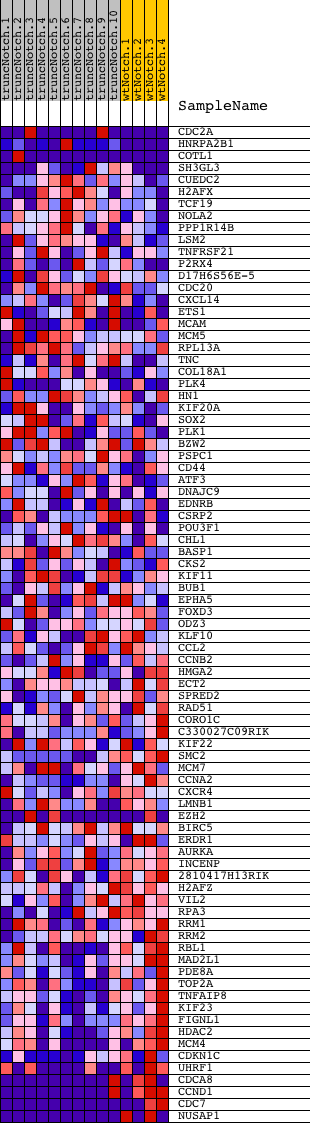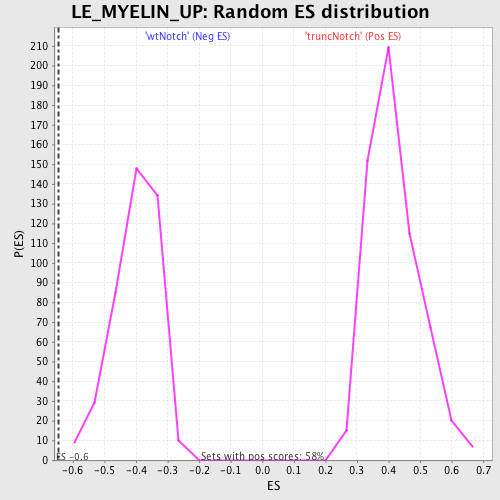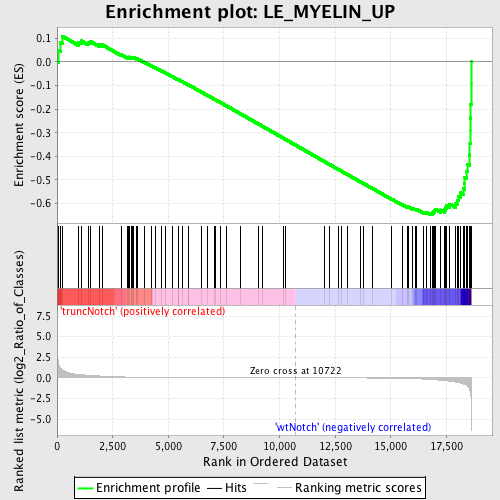
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | LE_MYELIN_UP |
| Enrichment Score (ES) | -0.64664525 |
| Normalized Enrichment Score (NES) | -1.6146641 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.17409864 |
| FWER p-Value | 0.934 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDC2A | 1570411 3800450 | 78 | 1.579 | 0.0475 | No | ||
| 2 | HNRPA2B1 | 1240619 6200161 6200494 6980408 | 152 | 1.154 | 0.0813 | No | ||
| 3 | COTL1 | 3840050 6350215 | 228 | 0.991 | 0.1097 | No | ||
| 4 | SH3GL3 | 3800112 | 957 | 0.416 | 0.0840 | No | ||
| 5 | CUEDC2 | 460095 | 1087 | 0.375 | 0.0893 | No | ||
| 6 | H2AFX | 3520082 | 1407 | 0.313 | 0.0823 | No | ||
| 7 | TCF19 | 6200152 | 1504 | 0.293 | 0.0868 | No | ||
| 8 | NOLA2 | 4060167 | 1903 | 0.230 | 0.0728 | No | ||
| 9 | PPP1R14B | 3800075 | 2036 | 0.211 | 0.0726 | No | ||
| 10 | LSM2 | 510082 | 2888 | 0.115 | 0.0304 | No | ||
| 11 | TNFRSF21 | 6380100 | 3175 | 0.096 | 0.0182 | No | ||
| 12 | P2RX4 | 2060497 6650162 | 3208 | 0.094 | 0.0195 | No | ||
| 13 | D17H6S56E-5 | 4760095 | 3234 | 0.092 | 0.0212 | No | ||
| 14 | CDC20 | 3440017 3440044 6220088 | 3342 | 0.085 | 0.0182 | No | ||
| 15 | CXCL14 | 840114 6450324 | 3395 | 0.082 | 0.0181 | No | ||
| 16 | ETS1 | 5270278 6450717 6620465 | 3446 | 0.079 | 0.0180 | No | ||
| 17 | MCAM | 5130270 6620577 | 3561 | 0.074 | 0.0142 | No | ||
| 18 | MCM5 | 2680647 | 3628 | 0.071 | 0.0130 | No | ||
| 19 | RPL13A | 2680519 5700142 | 3917 | 0.060 | -0.0006 | No | ||
| 20 | TNC | 670053 1780039 1980020 3060411 4780091 6860433 | 4234 | 0.050 | -0.0160 | No | ||
| 21 | COL18A1 | 610301 4570338 | 4405 | 0.046 | -0.0237 | No | ||
| 22 | PLK4 | 430162 5720110 | 4701 | 0.039 | -0.0383 | No | ||
| 23 | HN1 | 3360156 | 4859 | 0.036 | -0.0456 | No | ||
| 24 | KIF20A | 2650050 | 5168 | 0.031 | -0.0612 | No | ||
| 25 | SOX2 | 630487 2230491 | 5452 | 0.027 | -0.0756 | No | ||
| 26 | PLK1 | 1780369 2640121 | 5459 | 0.027 | -0.0750 | No | ||
| 27 | BZW2 | 940079 | 5640 | 0.025 | -0.0839 | No | ||
| 28 | PSPC1 | 1660017 | 5884 | 0.022 | -0.0963 | No | ||
| 29 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 5918 | 0.022 | -0.0974 | No | ||
| 30 | ATF3 | 1940546 | 6485 | 0.017 | -0.1274 | No | ||
| 31 | DNAJC9 | 6130019 | 6754 | 0.015 | -0.1414 | No | ||
| 32 | EDNRB | 4280717 6400435 | 7084 | 0.013 | -0.1587 | No | ||
| 33 | CSRP2 | 1030575 | 7128 | 0.012 | -0.1606 | No | ||
| 34 | POU3F1 | 3710022 | 7352 | 0.011 | -0.1723 | No | ||
| 35 | CHL1 | 50072 1500021 1580286 6660524 | 7597 | 0.010 | -0.1851 | No | ||
| 36 | BASP1 | 5700044 | 8224 | 0.008 | -0.2186 | No | ||
| 37 | CKS2 | 1410156 | 9073 | 0.005 | -0.2642 | No | ||
| 38 | KIF11 | 5390139 | 9250 | 0.004 | -0.2736 | No | ||
| 39 | BUB1 | 5390270 | 10161 | 0.002 | -0.3226 | No | ||
| 40 | EPHA5 | 2340671 | 10277 | 0.001 | -0.3288 | No | ||
| 41 | FOXD3 | 6550156 | 12005 | -0.004 | -0.4219 | No | ||
| 42 | ODZ3 | 3060725 6450292 | 12226 | -0.005 | -0.4336 | No | ||
| 43 | KLF10 | 4850056 | 12626 | -0.006 | -0.4549 | No | ||
| 44 | CCL2 | 4760019 | 12771 | -0.007 | -0.4624 | No | ||
| 45 | CCNB2 | 6510528 | 13033 | -0.009 | -0.4762 | No | ||
| 46 | HMGA2 | 2940121 3390647 5130279 6400136 | 13652 | -0.013 | -0.5092 | No | ||
| 47 | ECT2 | 3830086 | 13756 | -0.014 | -0.5143 | No | ||
| 48 | SPRED2 | 510138 540195 | 14186 | -0.019 | -0.5368 | No | ||
| 49 | RAD51 | 6110450 6980280 | 15032 | -0.039 | -0.5811 | No | ||
| 50 | CORO1C | 4570397 | 15504 | -0.061 | -0.6045 | No | ||
| 51 | C330027C09RIK | 2680450 2680685 | 15752 | -0.077 | -0.6153 | No | ||
| 52 | KIF22 | 1190368 | 15776 | -0.079 | -0.6140 | No | ||
| 53 | SMC2 | 4810133 | 15987 | -0.095 | -0.6222 | No | ||
| 54 | MCM7 | 3290292 5220056 | 16086 | -0.103 | -0.6241 | No | ||
| 55 | CCNA2 | 5290075 | 16158 | -0.110 | -0.6244 | No | ||
| 56 | CXCR4 | 4590519 | 16470 | -0.146 | -0.6364 | No | ||
| 57 | LMNB1 | 5890292 6020008 | 16609 | -0.167 | -0.6383 | No | ||
| 58 | EZH2 | 6130605 6380524 | 16764 | -0.189 | -0.6405 | Yes | ||
| 59 | BIRC5 | 110408 580014 1770632 | 16855 | -0.207 | -0.6385 | Yes | ||
| 60 | ERDR1 | 5890184 6370142 | 16900 | -0.215 | -0.6339 | Yes | ||
| 61 | AURKA | 780537 | 16964 | -0.225 | -0.6299 | Yes | ||
| 62 | INCENP | 520593 | 16986 | -0.229 | -0.6235 | Yes | ||
| 63 | 2810417H13RIK | 4850091 | 17233 | -0.283 | -0.6276 | Yes | ||
| 64 | H2AFZ | 1470168 | 17407 | -0.332 | -0.6260 | Yes | ||
| 65 | VIL2 | 5570161 | 17463 | -0.347 | -0.6176 | Yes | ||
| 66 | RPA3 | 5700136 | 17503 | -0.356 | -0.6081 | Yes | ||
| 67 | RRM1 | 4150433 | 17644 | -0.408 | -0.6023 | Yes | ||
| 68 | RRM2 | 6350059 6940162 | 17909 | -0.510 | -0.5999 | Yes | ||
| 69 | RBL1 | 3130372 | 17978 | -0.543 | -0.5858 | Yes | ||
| 70 | MAD2L1 | 4480725 | 18047 | -0.583 | -0.5704 | Yes | ||
| 71 | PDE8A | 5080446 | 18120 | -0.624 | -0.5539 | Yes | ||
| 72 | TOP2A | 360717 | 18276 | -0.748 | -0.5378 | Yes | ||
| 73 | TNFAIP8 | 2760551 | 18307 | -0.778 | -0.5139 | Yes | ||
| 74 | KIF23 | 5570112 | 18328 | -0.792 | -0.4891 | Yes | ||
| 75 | FIGNL1 | 6860390 | 18416 | -0.914 | -0.4639 | Yes | ||
| 76 | HDAC2 | 4050433 | 18455 | -0.999 | -0.4332 | Yes | ||
| 77 | MCM4 | 2760673 5420711 | 18538 | -1.324 | -0.3943 | Yes | ||
| 78 | CDKN1C | 6520577 | 18557 | -1.527 | -0.3453 | Yes | ||
| 79 | UHRF1 | 1410273 1660242 | 18573 | -1.658 | -0.2919 | Yes | ||
| 80 | CDCA8 | 2340286 6980019 | 18575 | -1.667 | -0.2374 | Yes | ||
| 81 | CCND1 | 460524 770309 3120576 6980398 | 18589 | -1.801 | -0.1792 | Yes | ||
| 82 | CDC7 | 4060546 4850041 | 18609 | -2.689 | -0.0922 | Yes | ||
| 83 | NUSAP1 | 940048 3120435 | 18611 | -2.829 | 0.0003 | Yes |