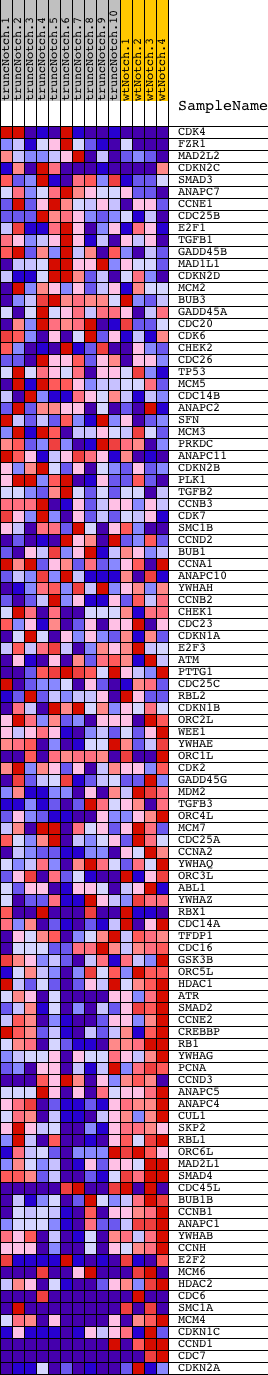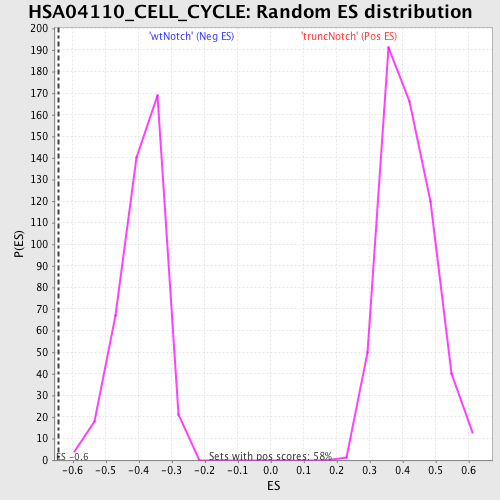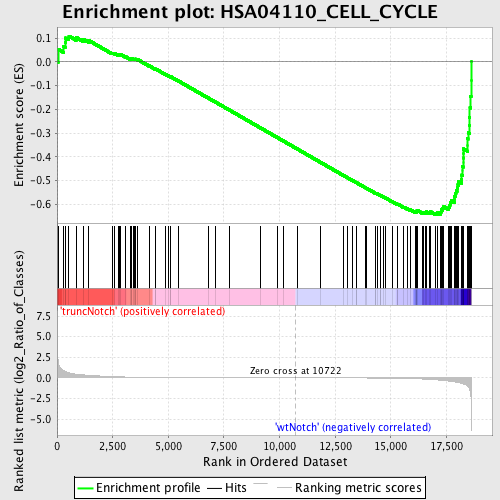
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | HSA04110_CELL_CYCLE |
| Enrichment Score (ES) | -0.6436782 |
| Normalized Enrichment Score (NES) | -1.6531919 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14653723 |
| FWER p-Value | 0.776 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDK4 | 540075 4540600 | 42 | 2.101 | 0.0536 | No | ||
| 2 | FZR1 | 6220504 6660670 | 285 | 0.915 | 0.0648 | No | ||
| 3 | MAD2L2 | 1240358 | 362 | 0.781 | 0.0815 | No | ||
| 4 | CDKN2C | 5050750 5130148 | 376 | 0.763 | 0.1011 | No | ||
| 5 | SMAD3 | 6450671 | 532 | 0.607 | 0.1089 | No | ||
| 6 | ANAPC7 | 5260274 | 854 | 0.450 | 0.1035 | No | ||
| 7 | CCNE1 | 110064 | 1182 | 0.356 | 0.0953 | No | ||
| 8 | CDC25B | 6940102 | 1428 | 0.308 | 0.0902 | No | ||
| 9 | E2F1 | 5360093 | 2480 | 0.155 | 0.0376 | No | ||
| 10 | TGFB1 | 1940162 | 2566 | 0.146 | 0.0369 | No | ||
| 11 | GADD45B | 2350408 | 2740 | 0.128 | 0.0310 | No | ||
| 12 | MAD1L1 | 5700035 | 2810 | 0.122 | 0.0305 | No | ||
| 13 | CDKN2D | 6040035 | 2854 | 0.118 | 0.0313 | No | ||
| 14 | MCM2 | 5050139 | 3062 | 0.103 | 0.0228 | No | ||
| 15 | BUB3 | 3170546 | 3320 | 0.087 | 0.0113 | No | ||
| 16 | GADD45A | 2900717 | 3324 | 0.086 | 0.0134 | No | ||
| 17 | CDC20 | 3440017 3440044 6220088 | 3342 | 0.085 | 0.0147 | No | ||
| 18 | CDK6 | 4920253 | 3448 | 0.079 | 0.0112 | No | ||
| 19 | CHEK2 | 610139 1050022 | 3468 | 0.079 | 0.0122 | No | ||
| 20 | CDC26 | 5570279 | 3537 | 0.075 | 0.0106 | No | ||
| 21 | TP53 | 6130707 | 3540 | 0.075 | 0.0124 | No | ||
| 22 | MCM5 | 2680647 | 3628 | 0.071 | 0.0096 | No | ||
| 23 | CDC14B | 2320497 3360162 | 4133 | 0.053 | -0.0162 | No | ||
| 24 | ANAPC2 | 6660438 | 4410 | 0.046 | -0.0299 | No | ||
| 25 | SFN | 6290301 7510608 | 4428 | 0.045 | -0.0296 | No | ||
| 26 | MCM3 | 5570068 | 4864 | 0.036 | -0.0521 | No | ||
| 27 | PRKDC | 1400072 4230541 | 5004 | 0.034 | -0.0587 | No | ||
| 28 | ANAPC11 | 1780601 | 5099 | 0.032 | -0.0630 | No | ||
| 29 | CDKN2B | 6020040 | 5117 | 0.032 | -0.0630 | No | ||
| 30 | PLK1 | 1780369 2640121 | 5459 | 0.027 | -0.0807 | No | ||
| 31 | TGFB2 | 4920292 | 6784 | 0.015 | -0.1519 | No | ||
| 32 | CCNB3 | 3360600 | 7115 | 0.013 | -0.1694 | No | ||
| 33 | CDK7 | 2640451 | 7752 | 0.009 | -0.2035 | No | ||
| 34 | SMC1B | 2450397 | 9159 | 0.004 | -0.2793 | No | ||
| 35 | CCND2 | 5340167 | 9902 | 0.002 | -0.3193 | No | ||
| 36 | BUB1 | 5390270 | 10161 | 0.002 | -0.3332 | No | ||
| 37 | CCNA1 | 6290113 | 10792 | -0.000 | -0.3672 | No | ||
| 38 | ANAPC10 | 870086 1170037 2260129 | 11836 | -0.003 | -0.4235 | No | ||
| 39 | YWHAH | 1660133 2810053 | 12889 | -0.008 | -0.4801 | No | ||
| 40 | CCNB2 | 6510528 | 13033 | -0.009 | -0.4876 | No | ||
| 41 | CHEK1 | 50167 1940300 | 13273 | -0.010 | -0.5002 | No | ||
| 42 | CDC23 | 3190593 | 13446 | -0.011 | -0.5092 | No | ||
| 43 | CDKN1A | 4050088 6400706 | 13471 | -0.011 | -0.5102 | No | ||
| 44 | E2F3 | 50162 460180 | 13841 | -0.015 | -0.5298 | No | ||
| 45 | ATM | 3610110 4050524 | 13898 | -0.015 | -0.5324 | No | ||
| 46 | PTTG1 | 520463 | 14307 | -0.021 | -0.5539 | No | ||
| 47 | CDC25C | 2570673 4760161 6520707 | 14329 | -0.021 | -0.5544 | No | ||
| 48 | RBL2 | 580446 1400670 | 14381 | -0.022 | -0.5566 | No | ||
| 49 | CDKN1B | 3800025 6450044 | 14515 | -0.024 | -0.5631 | No | ||
| 50 | ORC2L | 1990470 6510019 | 14531 | -0.025 | -0.5633 | No | ||
| 51 | WEE1 | 3390070 | 14671 | -0.027 | -0.5701 | No | ||
| 52 | YWHAE | 5310435 | 14779 | -0.030 | -0.5750 | No | ||
| 53 | ORC1L | 2370328 6110390 | 15055 | -0.039 | -0.5888 | No | ||
| 54 | CDK2 | 130484 2260301 4010088 5050110 | 15285 | -0.049 | -0.5999 | No | ||
| 55 | GADD45G | 2510142 | 15301 | -0.049 | -0.5994 | No | ||
| 56 | MDM2 | 3450053 5080138 | 15571 | -0.065 | -0.6122 | No | ||
| 57 | TGFB3 | 1070041 | 15747 | -0.077 | -0.6196 | No | ||
| 58 | ORC4L | 4230538 5550288 | 15863 | -0.085 | -0.6236 | No | ||
| 59 | MCM7 | 3290292 5220056 | 16086 | -0.103 | -0.6328 | No | ||
| 60 | CDC25A | 3800184 | 16131 | -0.107 | -0.6324 | No | ||
| 61 | CCNA2 | 5290075 | 16158 | -0.110 | -0.6308 | No | ||
| 62 | YWHAQ | 6760524 | 16174 | -0.112 | -0.6287 | No | ||
| 63 | ORC3L | 3060348 | 16182 | -0.113 | -0.6260 | No | ||
| 64 | ABL1 | 1050593 2030050 4010114 | 16410 | -0.138 | -0.6347 | No | ||
| 65 | YWHAZ | 1230717 | 16461 | -0.145 | -0.6335 | No | ||
| 66 | RBX1 | 2120010 2340047 | 16567 | -0.160 | -0.6349 | No | ||
| 67 | CDC14A | 4050132 | 16605 | -0.167 | -0.6325 | No | ||
| 68 | TFDP1 | 1980112 | 16756 | -0.187 | -0.6356 | No | ||
| 69 | CDC16 | 1940706 | 16789 | -0.194 | -0.6322 | No | ||
| 70 | GSK3B | 5360348 | 17003 | -0.232 | -0.6375 | Yes | ||
| 71 | ORC5L | 1940133 1940711 | 17097 | -0.252 | -0.6358 | Yes | ||
| 72 | HDAC1 | 2850670 | 17220 | -0.280 | -0.6350 | Yes | ||
| 73 | ATR | 6860273 | 17259 | -0.292 | -0.6293 | Yes | ||
| 74 | SMAD2 | 4200592 | 17271 | -0.295 | -0.6220 | Yes | ||
| 75 | CCNE2 | 3120537 | 17335 | -0.315 | -0.6171 | Yes | ||
| 76 | CREBBP | 5690035 7040050 | 17381 | -0.327 | -0.6108 | Yes | ||
| 77 | RB1 | 5900338 | 17613 | -0.394 | -0.6128 | Yes | ||
| 78 | YWHAG | 3780341 | 17650 | -0.409 | -0.6039 | Yes | ||
| 79 | PCNA | 940754 | 17662 | -0.414 | -0.5935 | Yes | ||
| 80 | CCND3 | 380528 730500 | 17704 | -0.426 | -0.5844 | Yes | ||
| 81 | ANAPC5 | 730164 7100685 | 17854 | -0.485 | -0.5795 | Yes | ||
| 82 | ANAPC4 | 4540338 | 17861 | -0.487 | -0.5669 | Yes | ||
| 83 | CUL1 | 1990632 | 17893 | -0.503 | -0.5552 | Yes | ||
| 84 | SKP2 | 360711 380093 4810368 | 17931 | -0.521 | -0.5433 | Yes | ||
| 85 | RBL1 | 3130372 | 17978 | -0.543 | -0.5314 | Yes | ||
| 86 | ORC6L | 2260592 | 18016 | -0.564 | -0.5184 | Yes | ||
| 87 | MAD2L1 | 4480725 | 18047 | -0.583 | -0.5045 | Yes | ||
| 88 | SMAD4 | 5670519 | 18179 | -0.658 | -0.4941 | Yes | ||
| 89 | CDC45L | 70537 3130114 | 18193 | -0.667 | -0.4770 | Yes | ||
| 90 | BUB1B | 1450288 | 18215 | -0.685 | -0.4600 | Yes | ||
| 91 | CCNB1 | 4590433 4780372 | 18216 | -0.685 | -0.4418 | Yes | ||
| 92 | ANAPC1 | 6350162 | 18250 | -0.719 | -0.4244 | Yes | ||
| 93 | YWHAB | 1740176 | 18251 | -0.719 | -0.4053 | Yes | ||
| 94 | CCNH | 3190347 | 18256 | -0.727 | -0.3862 | Yes | ||
| 95 | E2F2 | 5270609 5570377 7000465 | 18260 | -0.731 | -0.3669 | Yes | ||
| 96 | MCM6 | 60092 540181 6510110 | 18453 | -0.990 | -0.3510 | Yes | ||
| 97 | HDAC2 | 4050433 | 18455 | -0.999 | -0.3245 | Yes | ||
| 98 | CDC6 | 4570296 5360600 | 18471 | -1.030 | -0.2979 | Yes | ||
| 99 | SMC1A | 3060600 5700148 5890113 6370154 | 18529 | -1.261 | -0.2674 | Yes | ||
| 100 | MCM4 | 2760673 5420711 | 18538 | -1.324 | -0.2327 | Yes | ||
| 101 | CDKN1C | 6520577 | 18557 | -1.527 | -0.1930 | Yes | ||
| 102 | CCND1 | 460524 770309 3120576 6980398 | 18589 | -1.801 | -0.1468 | Yes | ||
| 103 | CDC7 | 4060546 4850041 | 18609 | -2.689 | -0.0764 | Yes | ||
| 104 | CDKN2A | 4670215 4760047 | 18612 | -2.884 | 0.0002 | Yes |