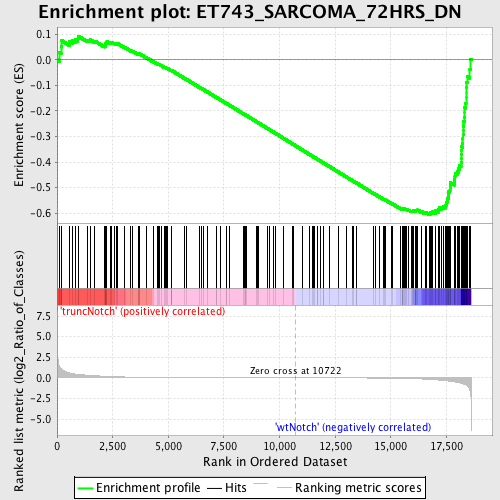
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #truncNotch_versus_wtNotch |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#truncNotch_versus_wtNotch |
| Upregulated in class | wtNotch |
| GeneSet | ET743_SARCOMA_72HRS_DN |
| Enrichment Score (ES) | -0.6049307 |
| Normalized Enrichment Score (NES) | -1.6466155 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15181804 |
| FWER p-Value | 0.822 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KIAA0859 | 510497 770722 1340687 6840711 | 103 | 1.447 | 0.0291 | No | ||
| 2 | SIRPA | 130465 5700332 | 178 | 1.090 | 0.0513 | No | ||
| 3 | WNT5B | 5080162 | 203 | 1.042 | 0.0750 | No | ||
| 4 | SLC2A1 | 2100609 | 545 | 0.599 | 0.0709 | No | ||
| 5 | GIPC1 | 4590348 5570438 5900707 | 702 | 0.507 | 0.0746 | No | ||
| 6 | HSPE1 | 3170438 | 818 | 0.461 | 0.0794 | No | ||
| 7 | TARDBP | 870390 2350093 | 938 | 0.419 | 0.0830 | No | ||
| 8 | LHFPL2 | 450020 | 948 | 0.418 | 0.0926 | No | ||
| 9 | BAG2 | 2900324 | 1373 | 0.320 | 0.0772 | No | ||
| 10 | RBM14 | 5340731 5690315 | 1486 | 0.297 | 0.0783 | No | ||
| 11 | SNIP1 | 5360092 | 1695 | 0.262 | 0.0733 | No | ||
| 12 | USP3 | 2060332 | 2126 | 0.198 | 0.0548 | No | ||
| 13 | FKBP1A | 2450368 | 2169 | 0.193 | 0.0571 | No | ||
| 14 | MID1IP1 | 2570739 | 2175 | 0.192 | 0.0615 | No | ||
| 15 | HSPA8 | 2120315 1050132 6550746 | 2184 | 0.191 | 0.0656 | No | ||
| 16 | ARFGAP1 | 4780364 5390358 | 2233 | 0.186 | 0.0675 | No | ||
| 17 | PANX1 | 6550632 | 2241 | 0.184 | 0.0715 | No | ||
| 18 | TOP1 | 770471 4060632 6650324 | 2420 | 0.162 | 0.0658 | No | ||
| 19 | SON | 3120148 5860239 7040403 | 2451 | 0.158 | 0.0679 | No | ||
| 20 | CHST2 | 3180682 | 2564 | 0.147 | 0.0654 | No | ||
| 21 | DNAJA2 | 2630253 6860358 | 2651 | 0.137 | 0.0640 | No | ||
| 22 | ZDHHC16 | 4230390 | 2702 | 0.131 | 0.0644 | No | ||
| 23 | DLG5 | 450215 | 3034 | 0.104 | 0.0490 | No | ||
| 24 | BUB3 | 3170546 | 3320 | 0.087 | 0.0356 | No | ||
| 25 | BLZF1 | 630088 | 3385 | 0.083 | 0.0341 | No | ||
| 26 | PDGFB | 3060440 6370008 | 3647 | 0.070 | 0.0217 | No | ||
| 27 | EPS15 | 4200215 | 3674 | 0.069 | 0.0219 | No | ||
| 28 | CLCN7 | 2970095 | 3689 | 0.069 | 0.0228 | No | ||
| 29 | EIF3S10 | 1190079 3840176 | 3700 | 0.068 | 0.0239 | No | ||
| 30 | ADSL | 5570484 | 4017 | 0.057 | 0.0081 | No | ||
| 31 | MRPL1 | 430626 3130129 5220152 | 4323 | 0.048 | -0.0072 | No | ||
| 32 | PSMA5 | 5390537 | 4502 | 0.044 | -0.0158 | No | ||
| 33 | FAS | 4480112 6110040 | 4574 | 0.042 | -0.0187 | No | ||
| 34 | RCC1 | 2480358 | 4585 | 0.042 | -0.0182 | No | ||
| 35 | IL1RL1 | 3990224 6020347 | 4680 | 0.040 | -0.0224 | No | ||
| 36 | THRAP3 | 6180309 | 4835 | 0.037 | -0.0298 | No | ||
| 37 | MCM3 | 5570068 | 4864 | 0.036 | -0.0305 | No | ||
| 38 | SEH1L | 4590563 | 4921 | 0.035 | -0.0327 | No | ||
| 39 | PAPPA | 4230463 | 4975 | 0.034 | -0.0347 | No | ||
| 40 | ANLN | 1780113 2470537 6840047 | 5124 | 0.032 | -0.0420 | No | ||
| 41 | MTMR4 | 4540524 | 5132 | 0.032 | -0.0416 | No | ||
| 42 | UBE2C | 6130017 | 5158 | 0.031 | -0.0422 | No | ||
| 43 | B4GALT5 | 1230692 | 5737 | 0.024 | -0.0730 | No | ||
| 44 | CTGF | 4540577 | 5795 | 0.023 | -0.0755 | No | ||
| 45 | NUFIP1 | 6350241 | 6390 | 0.017 | -0.1073 | No | ||
| 46 | HS3ST3B1 | 380040 | 6504 | 0.016 | -0.1130 | No | ||
| 47 | EIF4B | 5390563 5270577 | 6597 | 0.016 | -0.1176 | No | ||
| 48 | RNF14 | 50402 1340041 6370687 | 6752 | 0.015 | -0.1256 | No | ||
| 49 | HSPA1L | 4010538 | 7148 | 0.012 | -0.1467 | No | ||
| 50 | TIAM2 | 360195 | 7150 | 0.012 | -0.1465 | No | ||
| 51 | CPT1A | 6350093 | 7327 | 0.011 | -0.1557 | No | ||
| 52 | BIRC6 | 70148 | 7596 | 0.010 | -0.1700 | No | ||
| 53 | CDH11 | 1230133 2100180 4610110 | 7630 | 0.010 | -0.1716 | No | ||
| 54 | CDK7 | 2640451 | 7752 | 0.009 | -0.1779 | No | ||
| 55 | IPO7 | 2190746 | 8359 | 0.007 | -0.2106 | No | ||
| 56 | CXXC5 | 840010 | 8418 | 0.007 | -0.2136 | No | ||
| 57 | RAB5A | 6520324 | 8483 | 0.007 | -0.2169 | No | ||
| 58 | JAG1 | 3440390 | 8486 | 0.007 | -0.2168 | No | ||
| 59 | ARHGEF2 | 3360577 | 8534 | 0.006 | -0.2192 | No | ||
| 60 | CRSP6 | 2940358 | 8963 | 0.005 | -0.2423 | No | ||
| 61 | FOSL1 | 430021 | 8996 | 0.005 | -0.2439 | No | ||
| 62 | EPS8 | 7050204 | 9067 | 0.005 | -0.2476 | No | ||
| 63 | ACSL3 | 3140195 | 9470 | 0.004 | -0.2693 | No | ||
| 64 | FZD1 | 3140215 | 9544 | 0.003 | -0.2732 | No | ||
| 65 | XRCC5 | 7100286 | 9727 | 0.003 | -0.2830 | No | ||
| 66 | MFAP1 | 6660092 | 9816 | 0.003 | -0.2877 | No | ||
| 67 | BUB1 | 5390270 | 10161 | 0.002 | -0.3063 | No | ||
| 68 | MRPL35 | 1990671 | 10578 | 0.000 | -0.3288 | No | ||
| 69 | CFLAR | 2320605 6180440 | 10607 | 0.000 | -0.3303 | No | ||
| 70 | ATG5 | 6200433 5360324 | 11031 | -0.001 | -0.3532 | No | ||
| 71 | FBXL7 | 6620711 | 11360 | -0.002 | -0.3710 | No | ||
| 72 | NCOA6 | 1780333 6450110 | 11459 | -0.002 | -0.3762 | No | ||
| 73 | GDI2 | 2370068 | 11545 | -0.002 | -0.3808 | No | ||
| 74 | VEGFC | 5910494 | 11570 | -0.002 | -0.3820 | No | ||
| 75 | TDG | 2900358 2940079 | 11705 | -0.003 | -0.3892 | No | ||
| 76 | PLAG1 | 1450142 3870139 6520039 | 11708 | -0.003 | -0.3893 | No | ||
| 77 | SPRY2 | 5860184 | 11837 | -0.003 | -0.3961 | No | ||
| 78 | GGPS1 | 840128 2320215 | 11953 | -0.004 | -0.4023 | No | ||
| 79 | CRK | 1230162 4780128 | 11959 | -0.004 | -0.4024 | No | ||
| 80 | MYC | 380541 4670170 | 12225 | -0.005 | -0.4167 | No | ||
| 81 | MTIF2 | 2260270 | 12628 | -0.006 | -0.4383 | No | ||
| 82 | LYN | 6040600 | 12993 | -0.008 | -0.4579 | No | ||
| 83 | TCERG1 | 2350551 4540397 5220746 | 13257 | -0.010 | -0.4719 | No | ||
| 84 | PRKD3 | 5220520 5890519 | 13330 | -0.010 | -0.4755 | No | ||
| 85 | CDC23 | 3190593 | 13446 | -0.011 | -0.4815 | No | ||
| 86 | RBM27 | 4050603 | 13460 | -0.011 | -0.4819 | No | ||
| 87 | SFRS9 | 3800047 | 14204 | -0.019 | -0.5217 | No | ||
| 88 | CDC25C | 2570673 4760161 6520707 | 14329 | -0.021 | -0.5279 | No | ||
| 89 | SSFA2 | 3870133 | 14488 | -0.024 | -0.5359 | No | ||
| 90 | WEE1 | 3390070 | 14671 | -0.027 | -0.5451 | No | ||
| 91 | GOPC | 6620400 | 14714 | -0.029 | -0.5467 | No | ||
| 92 | CDC5L | 3130446 3390025 6400204 | 14776 | -0.030 | -0.5493 | No | ||
| 93 | USP12 | 6130735 6620280 | 15014 | -0.038 | -0.5612 | No | ||
| 94 | FNDC3A | 2690097 3290044 | 15085 | -0.040 | -0.5641 | No | ||
| 95 | PSMB1 | 2940402 | 15414 | -0.055 | -0.5805 | No | ||
| 96 | CORO1C | 4570397 | 15504 | -0.061 | -0.5839 | No | ||
| 97 | SNRPG | 2340551 | 15505 | -0.061 | -0.5824 | No | ||
| 98 | PPP1CC | 6380300 2510647 | 15544 | -0.063 | -0.5830 | No | ||
| 99 | MRPL15 | 360390 | 15566 | -0.065 | -0.5825 | No | ||
| 100 | FBXL3 | 5910142 | 15606 | -0.067 | -0.5831 | No | ||
| 101 | SMARCA5 | 6620050 | 15678 | -0.072 | -0.5852 | No | ||
| 102 | GCNT1 | 940129 1780215 2370253 | 15697 | -0.073 | -0.5844 | No | ||
| 103 | JMJD2B | 6350372 6400471 | 15794 | -0.080 | -0.5877 | No | ||
| 104 | ARFGEF2 | 5550500 | 15917 | -0.089 | -0.5922 | No | ||
| 105 | GABPB2 | 6130450 7050735 | 15964 | -0.093 | -0.5924 | No | ||
| 106 | ERCC3 | 6900008 | 15992 | -0.095 | -0.5916 | No | ||
| 107 | PSMC6 | 2810021 | 16015 | -0.097 | -0.5904 | No | ||
| 108 | ZDHHC5 | 1690711 | 16093 | -0.104 | -0.5921 | No | ||
| 109 | CYHR1 | 940609 2810463 3190332 3290692 4280411 | 16123 | -0.106 | -0.5912 | No | ||
| 110 | RPS7 | 6860113 | 16129 | -0.107 | -0.5889 | No | ||
| 111 | CCNA2 | 5290075 | 16158 | -0.110 | -0.5877 | No | ||
| 112 | PRIM2A | 7000692 | 16201 | -0.114 | -0.5873 | No | ||
| 113 | BIRC2 | 2940435 4730020 | 16364 | -0.132 | -0.5929 | No | ||
| 114 | FADD | 2360593 4210692 6520019 | 16548 | -0.158 | -0.5990 | No | ||
| 115 | EML4 | 5700373 | 16596 | -0.165 | -0.5976 | No | ||
| 116 | STK17B | 130091 | 16732 | -0.184 | -0.6005 | Yes | ||
| 117 | TXNRD1 | 6590446 | 16784 | -0.194 | -0.5986 | Yes | ||
| 118 | SUPV3L1 | 1170072 | 16847 | -0.206 | -0.5971 | Yes | ||
| 119 | SIRT1 | 1190731 | 16889 | -0.212 | -0.5942 | Yes | ||
| 120 | MAT2A | 4070026 4730079 6020280 | 17019 | -0.235 | -0.5955 | Yes | ||
| 121 | GTF2B | 6860685 | 17024 | -0.236 | -0.5901 | Yes | ||
| 122 | GNL2 | 6620022 | 17124 | -0.257 | -0.5893 | Yes | ||
| 123 | MAPK14 | 5290731 | 17146 | -0.262 | -0.5841 | Yes | ||
| 124 | TMPO | 4050494 | 17165 | -0.266 | -0.5787 | Yes | ||
| 125 | ELOVL6 | 5340746 | 17264 | -0.293 | -0.5770 | Yes | ||
| 126 | BCL2L1 | 1580452 4200152 5420484 | 17359 | -0.321 | -0.5744 | Yes | ||
| 127 | TNFSF5IP1 | 2060095 | 17457 | -0.346 | -0.5714 | Yes | ||
| 128 | MAP3K7 | 6040068 | 17481 | -0.352 | -0.5642 | Yes | ||
| 129 | PPP2CB | 5570593 | 17505 | -0.356 | -0.5569 | Yes | ||
| 130 | PSMD12 | 730044 | 17531 | -0.365 | -0.5495 | Yes | ||
| 131 | PJA2 | 430537 | 17555 | -0.371 | -0.5418 | Yes | ||
| 132 | UBE2D3 | 3190452 | 17584 | -0.381 | -0.5342 | Yes | ||
| 133 | GTF2H1 | 2570746 2650148 | 17587 | -0.381 | -0.5252 | Yes | ||
| 134 | POLD3 | 6400278 | 17603 | -0.388 | -0.5167 | Yes | ||
| 135 | FNTA | 70059 | 17657 | -0.412 | -0.5097 | Yes | ||
| 136 | PCNA | 940754 | 17662 | -0.414 | -0.4999 | Yes | ||
| 137 | RIPK1 | 3190025 | 17668 | -0.416 | -0.4902 | Yes | ||
| 138 | SLC25A17 | 2100369 | 17674 | -0.418 | -0.4805 | Yes | ||
| 139 | PIK3C3 | 6590717 | 17848 | -0.483 | -0.4783 | Yes | ||
| 140 | UBE2D2 | 4590671 6510446 | 17859 | -0.486 | -0.4671 | Yes | ||
| 141 | DDX20 | 3290348 | 17882 | -0.495 | -0.4565 | Yes | ||
| 142 | EXOC2 | 3190132 | 17889 | -0.499 | -0.4448 | Yes | ||
| 143 | RAB1A | 2370671 | 17979 | -0.543 | -0.4366 | Yes | ||
| 144 | PSMA4 | 4560427 | 18051 | -0.585 | -0.4264 | Yes | ||
| 145 | XBP1 | 3840594 | 18082 | -0.605 | -0.4135 | Yes | ||
| 146 | RABGGTB | 3120037 | 18157 | -0.647 | -0.4020 | Yes | ||
| 147 | OGT | 2360131 4610333 | 18167 | -0.653 | -0.3868 | Yes | ||
| 148 | RAP1B | 3710138 | 18172 | -0.654 | -0.3713 | Yes | ||
| 149 | SMAD4 | 5670519 | 18179 | -0.658 | -0.3559 | Yes | ||
| 150 | EMP1 | 4120438 | 18184 | -0.662 | -0.3402 | Yes | ||
| 151 | PPP2R5E | 70671 2120056 | 18221 | -0.689 | -0.3256 | Yes | ||
| 152 | MCTS1 | 6130133 | 18240 | -0.706 | -0.3097 | Yes | ||
| 153 | HSPH1 | 1690017 | 18244 | -0.711 | -0.2928 | Yes | ||
| 154 | CCNH | 3190347 | 18256 | -0.727 | -0.2759 | Yes | ||
| 155 | CCT4 | 6380161 | 18262 | -0.731 | -0.2587 | Yes | ||
| 156 | TOP2A | 360717 | 18276 | -0.748 | -0.2414 | Yes | ||
| 157 | SRPK2 | 6380341 | 18312 | -0.780 | -0.2246 | Yes | ||
| 158 | DDX21 | 6100446 | 18324 | -0.788 | -0.2063 | Yes | ||
| 159 | GTF2H2 | 5130358 | 18326 | -0.788 | -0.1874 | Yes | ||
| 160 | SLC4A1AP | 3190576 | 18363 | -0.843 | -0.1691 | Yes | ||
| 161 | RBM17 | 870592 | 18382 | -0.867 | -0.1493 | Yes | ||
| 162 | RABGAP1 | 5050397 | 18384 | -0.869 | -0.1285 | Yes | ||
| 163 | TBK1 | 5690338 | 18385 | -0.870 | -0.1076 | Yes | ||
| 164 | LBR | 4540671 | 18390 | -0.872 | -0.0869 | Yes | ||
| 165 | HDAC2 | 4050433 | 18455 | -0.999 | -0.0664 | Yes | ||
| 166 | ATP2A2 | 1090075 3990279 | 18554 | -1.475 | -0.0363 | Yes | ||
| 167 | ACTL6A | 5220543 6290541 2450093 4590332 5270722 | 18570 | -1.652 | 0.0025 | Yes |

