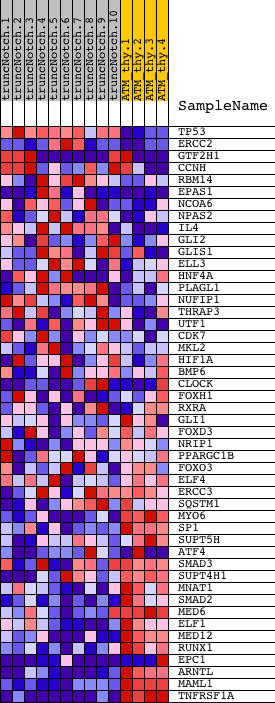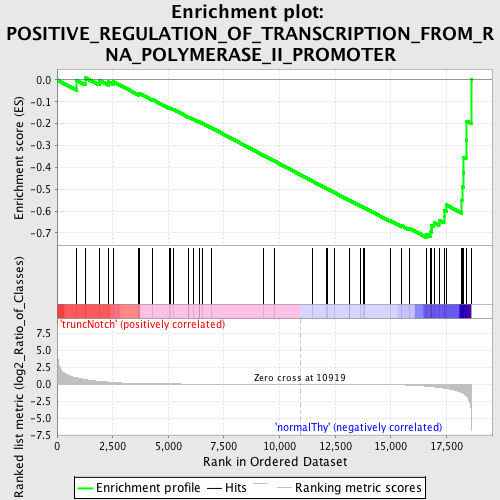
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | POSITIVE_REGULATION_OF_TRANSCRIPTION_FROM_RNA_POLYMERASE_II_PROMOTER |
| Enrichment Score (ES) | -0.72096044 |
| Normalized Enrichment Score (NES) | -1.5811186 |
| Nominal p-value | 0.002631579 |
| FDR q-value | 0.6392681 |
| FWER p-Value | 0.846 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | 6130707 | 880 | 0.930 | -0.0018 | No | ||
| 2 | ERCC2 | 2360750 4060390 6550138 | 1264 | 0.677 | 0.0107 | No | ||
| 3 | GTF2H1 | 2570746 2650148 | 1894 | 0.430 | -0.0021 | No | ||
| 4 | CCNH | 3190347 | 2311 | 0.315 | -0.0091 | No | ||
| 5 | RBM14 | 5340731 5690315 | 2517 | 0.260 | -0.0074 | No | ||
| 6 | EPAS1 | 5290156 | 3671 | 0.095 | -0.0648 | No | ||
| 7 | NCOA6 | 1780333 6450110 | 3693 | 0.093 | -0.0614 | No | ||
| 8 | NPAS2 | 770670 | 4268 | 0.064 | -0.0892 | No | ||
| 9 | IL4 | 6020537 | 5032 | 0.041 | -0.1283 | No | ||
| 10 | GLI2 | 3060632 | 5094 | 0.039 | -0.1296 | No | ||
| 11 | GLIS1 | 840471 | 5216 | 0.037 | -0.1343 | No | ||
| 12 | ELL3 | 1770484 | 5897 | 0.025 | -0.1697 | No | ||
| 13 | HNF4A | 6620440 | 5926 | 0.025 | -0.1700 | No | ||
| 14 | PLAGL1 | 3190082 6200193 | 6141 | 0.022 | -0.1805 | No | ||
| 15 | NUFIP1 | 6350241 | 6384 | 0.019 | -0.1926 | No | ||
| 16 | THRAP3 | 6180309 | 6393 | 0.019 | -0.1920 | No | ||
| 17 | UTF1 | 5700593 | 6516 | 0.018 | -0.1977 | No | ||
| 18 | CDK7 | 2640451 | 6958 | 0.015 | -0.2207 | No | ||
| 19 | MKL2 | 5700204 | 9263 | 0.005 | -0.3446 | No | ||
| 20 | HIF1A | 5670605 | 9787 | 0.003 | -0.3726 | No | ||
| 21 | BMP6 | 2100128 | 11500 | -0.002 | -0.4647 | No | ||
| 22 | CLOCK | 1410070 2940292 3390075 | 12130 | -0.004 | -0.4984 | No | ||
| 23 | FOXH1 | 2060180 | 12169 | -0.004 | -0.5002 | No | ||
| 24 | RXRA | 3800471 | 12485 | -0.005 | -0.5169 | No | ||
| 25 | GLI1 | 4560176 | 13129 | -0.009 | -0.5511 | No | ||
| 26 | FOXD3 | 6550156 | 13160 | -0.009 | -0.5523 | No | ||
| 27 | NRIP1 | 3190039 | 13626 | -0.014 | -0.5766 | No | ||
| 28 | PPARGC1B | 3290114 | 13767 | -0.015 | -0.5834 | No | ||
| 29 | FOXO3 | 2510484 4480451 | 13812 | -0.016 | -0.5850 | No | ||
| 30 | ELF4 | 4850193 | 14976 | -0.056 | -0.6449 | No | ||
| 31 | ERCC3 | 6900008 | 15501 | -0.101 | -0.6682 | No | ||
| 32 | SQSTM1 | 6550056 | 15821 | -0.137 | -0.6787 | No | ||
| 33 | MYO6 | 2190332 | 16607 | -0.296 | -0.7065 | Yes | ||
| 34 | SP1 | 6590017 | 16789 | -0.347 | -0.6992 | Yes | ||
| 35 | SUPT5H | 4150546 | 16813 | -0.353 | -0.6832 | Yes | ||
| 36 | ATF4 | 730441 1740195 | 16819 | -0.355 | -0.6661 | Yes | ||
| 37 | SMAD3 | 6450671 | 16940 | -0.392 | -0.6533 | Yes | ||
| 38 | SUPT4H1 | 6200056 | 17166 | -0.478 | -0.6420 | Yes | ||
| 39 | MNAT1 | 730292 2350364 | 17391 | -0.575 | -0.6259 | Yes | ||
| 40 | SMAD2 | 4200592 | 17397 | -0.576 | -0.5980 | Yes | ||
| 41 | MED6 | 6840500 | 17488 | -0.626 | -0.5722 | Yes | ||
| 42 | ELF1 | 4780450 | 18178 | -1.217 | -0.5496 | Yes | ||
| 43 | MED12 | 4920095 | 18202 | -1.264 | -0.4889 | Yes | ||
| 44 | RUNX1 | 3840711 | 18253 | -1.370 | -0.4245 | Yes | ||
| 45 | EPC1 | 1570050 5290095 6900193 | 18284 | -1.437 | -0.3558 | Yes | ||
| 46 | ARNTL | 3170463 | 18381 | -1.727 | -0.2763 | Yes | ||
| 47 | MAML1 | 2760008 | 18415 | -1.816 | -0.1891 | Yes | ||
| 48 | TNFRSF1A | 1090390 6520735 | 18609 | -4.080 | 0.0004 | Yes |