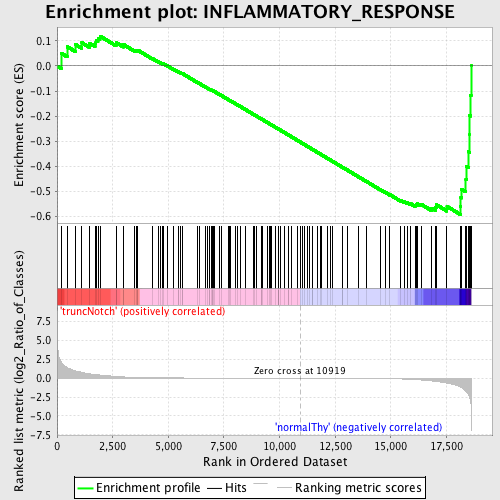
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.59381497 |
| Normalized Enrichment Score (NES) | -1.4880551 |
| Nominal p-value | 0.0057306592 |
| FDR q-value | 0.60365415 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 2970019 | 183 | 2.087 | 0.0512 | No | ||
| 2 | ABCF1 | 2630707 2760273 3520725 | 452 | 1.395 | 0.0776 | No | ||
| 3 | C2 | 5390465 | 826 | 0.971 | 0.0858 | No | ||
| 4 | CCL4 | 50368 430047 | 1111 | 0.774 | 0.0931 | No | ||
| 5 | CXCL1 | 2690537 | 1459 | 0.580 | 0.0913 | No | ||
| 6 | TGFB1 | 1940162 | 1706 | 0.484 | 0.0922 | No | ||
| 7 | ALOX5AP | 3170440 | 1747 | 0.472 | 0.1039 | No | ||
| 8 | BLNK | 70288 6590092 | 1838 | 0.443 | 0.1120 | No | ||
| 9 | NFKB1 | 5420358 | 1928 | 0.422 | 0.1195 | No | ||
| 10 | IL10RB | 2900025 | 2651 | 0.227 | 0.0871 | No | ||
| 11 | TPST1 | 2570358 | 2658 | 0.226 | 0.0934 | No | ||
| 12 | CCL3 | 2810092 | 2985 | 0.168 | 0.0807 | No | ||
| 13 | SCYE1 | 4730736 | 2993 | 0.167 | 0.0852 | No | ||
| 14 | AIF1 | 130332 5220017 | 3480 | 0.110 | 0.0622 | No | ||
| 15 | LTB4R | 1770056 | 3581 | 0.102 | 0.0598 | No | ||
| 16 | SCG2 | 5670438 130671 | 3599 | 0.100 | 0.0618 | No | ||
| 17 | CHST2 | 3180682 | 3616 | 0.099 | 0.0638 | No | ||
| 18 | IL13 | 7040021 | 4284 | 0.063 | 0.0296 | No | ||
| 19 | CXCL2 | 610398 | 4552 | 0.054 | 0.0167 | No | ||
| 20 | CCL24 | 670129 | 4658 | 0.051 | 0.0126 | No | ||
| 21 | LBP | 6860019 | 4755 | 0.048 | 0.0088 | No | ||
| 22 | CHRNA7 | 2970446 | 4779 | 0.047 | 0.0089 | No | ||
| 23 | AHSG | 6450014 | 4946 | 0.042 | 0.0012 | No | ||
| 24 | ELF3 | 770551 6180019 | 5236 | 0.036 | -0.0134 | No | ||
| 25 | AOAH | 5390672 | 5443 | 0.032 | -0.0236 | No | ||
| 26 | ADORA2A | 1990687 | 5457 | 0.032 | -0.0233 | No | ||
| 27 | GHSR | 1980465 7050521 | 5535 | 0.031 | -0.0266 | No | ||
| 28 | NFX1 | 450504 | 5632 | 0.029 | -0.0309 | No | ||
| 29 | IL5 | 6370364 1050193 | 5640 | 0.029 | -0.0305 | No | ||
| 30 | PLA2G7 | 4730092 | 6310 | 0.020 | -0.0660 | No | ||
| 31 | APCS | 2060500 | 6391 | 0.019 | -0.0698 | No | ||
| 32 | ORM2 | 2100048 | 6690 | 0.017 | -0.0854 | No | ||
| 33 | AOC3 | 6840129 | 6754 | 0.016 | -0.0883 | No | ||
| 34 | CCL22 | 6380086 | 6862 | 0.016 | -0.0936 | No | ||
| 35 | CRP | 3140711 | 6867 | 0.016 | -0.0934 | No | ||
| 36 | CCR1 | 3440672 | 6921 | 0.015 | -0.0958 | No | ||
| 37 | CCR3 | 50427 | 6972 | 0.015 | -0.0981 | No | ||
| 38 | PLA2G2D | 2850047 | 6991 | 0.015 | -0.0986 | No | ||
| 39 | KNG1 | 6400576 6770347 | 7036 | 0.014 | -0.1006 | No | ||
| 40 | EDG3 | 3360110 | 7092 | 0.014 | -0.1031 | No | ||
| 41 | CDO1 | 2480279 | 7301 | 0.013 | -0.1140 | No | ||
| 42 | FOS | 1850315 | 7390 | 0.012 | -0.1184 | No | ||
| 43 | CX3CL1 | 3990707 | 7694 | 0.011 | -0.1345 | No | ||
| 44 | ADORA1 | 6370520 | 7753 | 0.010 | -0.1373 | No | ||
| 45 | IL8RB | 450592 1170537 | 7790 | 0.010 | -0.1390 | No | ||
| 46 | IL1A | 3610056 | 8005 | 0.009 | -0.1502 | No | ||
| 47 | ORM1 | 1500563 | 8120 | 0.009 | -0.1562 | No | ||
| 48 | NMI | 520270 | 8238 | 0.008 | -0.1622 | No | ||
| 49 | SELE | 7100368 | 8486 | 0.007 | -0.1754 | No | ||
| 50 | HRH1 | 840100 | 8822 | 0.006 | -0.1933 | No | ||
| 51 | CCR5 | 1980072 5910722 | 8890 | 0.006 | -0.1967 | No | ||
| 52 | IL9 | 3440338 | 8982 | 0.006 | -0.2015 | No | ||
| 53 | IL8RA | 1580100 | 9181 | 0.005 | -0.2121 | No | ||
| 54 | CCL11 | 1770347 | 9186 | 0.005 | -0.2121 | No | ||
| 55 | C3AR1 | 5720131 | 9234 | 0.005 | -0.2145 | No | ||
| 56 | IRAK2 | 730400 | 9447 | 0.004 | -0.2259 | No | ||
| 57 | CXCL10 | 2450408 | 9533 | 0.004 | -0.2303 | No | ||
| 58 | ANXA1 | 2320053 | 9607 | 0.004 | -0.2342 | No | ||
| 59 | IL20 | 4850647 | 9632 | 0.004 | -0.2354 | No | ||
| 60 | PTX3 | 870309 3520102 | 9808 | 0.003 | -0.2447 | No | ||
| 61 | NLRP3 | 1980064 4010180 | 9931 | 0.003 | -0.2512 | No | ||
| 62 | XCR1 | 5080072 | 9942 | 0.003 | -0.2517 | No | ||
| 63 | NOX4 | 6040136 | 9957 | 0.003 | -0.2524 | No | ||
| 64 | F11R | 1740128 | 10053 | 0.002 | -0.2574 | No | ||
| 65 | AGER | 5860538 5910180 | 10211 | 0.002 | -0.2659 | No | ||
| 66 | ADORA3 | 630333 | 10395 | 0.002 | -0.2757 | No | ||
| 67 | PLA2G2E | 4060128 | 10410 | 0.001 | -0.2764 | No | ||
| 68 | IFNA2 | 4150162 | 10548 | 0.001 | -0.2838 | No | ||
| 69 | CYBB | 3870020 | 10809 | 0.000 | -0.2978 | No | ||
| 70 | IL1RAP | 50546 450711 1580397 3940301 5890181 6370373 | 10939 | -0.000 | -0.3048 | No | ||
| 71 | AOX1 | 110082 6290450 | 11009 | -0.000 | -0.3085 | No | ||
| 72 | IL17C | 5690347 | 11109 | -0.001 | -0.3139 | No | ||
| 73 | CXCL11 | 1090551 | 11236 | -0.001 | -0.3206 | No | ||
| 74 | CCR2 | 1450519 | 11357 | -0.001 | -0.3271 | No | ||
| 75 | MBL2 | 6370446 | 11483 | -0.002 | -0.3338 | No | ||
| 76 | MEFV | 510364 | 11493 | -0.002 | -0.3342 | No | ||
| 77 | TNFAIP6 | 3390161 | 11686 | -0.002 | -0.3445 | No | ||
| 78 | CD40LG | 3120270 | 11850 | -0.003 | -0.3533 | No | ||
| 79 | NFATC4 | 2470735 | 11894 | -0.003 | -0.3555 | No | ||
| 80 | ALOX15 | 6620333 | 12159 | -0.004 | -0.3696 | No | ||
| 81 | FPRL1 | 3360451 | 12275 | -0.004 | -0.3757 | No | ||
| 82 | TACR1 | 70358 3840411 | 12366 | -0.005 | -0.3804 | No | ||
| 83 | IL18RAP | 3850088 | 12831 | -0.007 | -0.4053 | No | ||
| 84 | LY75 | 1990358 4590136 | 12834 | -0.007 | -0.4052 | No | ||
| 85 | HDAC9 | 1990010 2260133 | 13034 | -0.008 | -0.4157 | No | ||
| 86 | CFHR1 | 1850050 5550519 | 13562 | -0.013 | -0.4438 | No | ||
| 87 | GHRL | 2360619 6760438 | 13918 | -0.017 | -0.4625 | No | ||
| 88 | HDAC5 | 2370035 6370139 | 14528 | -0.033 | -0.4944 | No | ||
| 89 | CCR4 | 4480184 | 14767 | -0.044 | -0.5060 | No | ||
| 90 | PARP4 | 4070095 | 14959 | -0.055 | -0.5147 | No | ||
| 91 | KLRG1 | 2360133 | 15435 | -0.094 | -0.5377 | No | ||
| 92 | CD40 | 2760471 2940390 6220402 6220452 | 15600 | -0.112 | -0.5432 | No | ||
| 93 | MGLL | 2030446 4210520 | 15738 | -0.128 | -0.5469 | No | ||
| 94 | CXCL9 | 1570673 | 15902 | -0.148 | -0.5514 | No | ||
| 95 | RAC1 | 4810687 | 16093 | -0.176 | -0.5565 | No | ||
| 96 | RIPK2 | 5050072 6290632 | 16153 | -0.187 | -0.5542 | No | ||
| 97 | NFRKB | 3130538 | 16188 | -0.195 | -0.5503 | No | ||
| 98 | VPS45 | 1770035 | 16356 | -0.228 | -0.5527 | No | ||
| 99 | RELA | 3830075 | 16834 | -0.358 | -0.5680 | Yes | ||
| 100 | PRDX5 | 1660592 2030091 | 17019 | -0.421 | -0.5656 | Yes | ||
| 101 | NOD1 | 2360368 | 17050 | -0.431 | -0.5546 | Yes | ||
| 102 | GPR68 | 1190537 | 17524 | -0.644 | -0.5613 | Yes | ||
| 103 | HDAC7A | 2570278 | 18126 | -1.153 | -0.5601 | Yes | ||
| 104 | CXCR4 | 4590519 | 18148 | -1.184 | -0.5266 | Yes | ||
| 105 | NFE2L1 | 6130053 | 18175 | -1.214 | -0.4924 | Yes | ||
| 106 | CD97 | 1090673 1690575 | 18375 | -1.717 | -0.4529 | Yes | ||
| 107 | S100A9 | 7050528 | 18408 | -1.792 | -0.4022 | Yes | ||
| 108 | S100A8 | 70112 | 18489 | -2.180 | -0.3427 | Yes | ||
| 109 | CCL5 | 3710397 | 18540 | -2.493 | -0.2725 | Yes | ||
| 110 | CCR7 | 2060687 | 18547 | -2.525 | -0.1989 | Yes | ||
| 111 | NFATC3 | 5050520 | 18574 | -2.838 | -0.1172 | Yes | ||
| 112 | TNFRSF1A | 1090390 6520735 | 18609 | -4.080 | 0.0004 | Yes |

