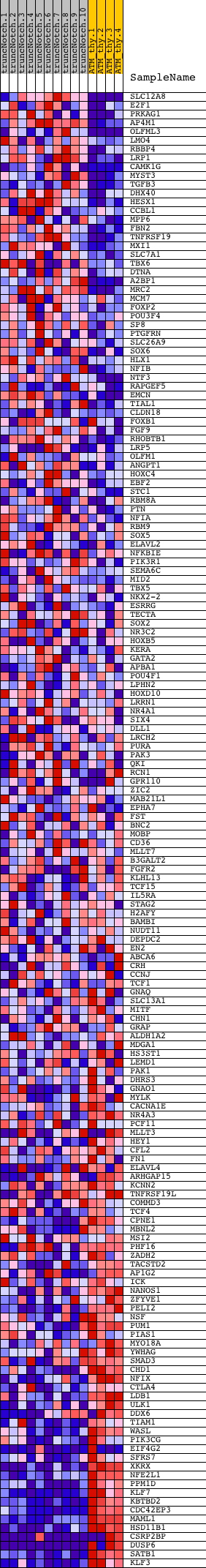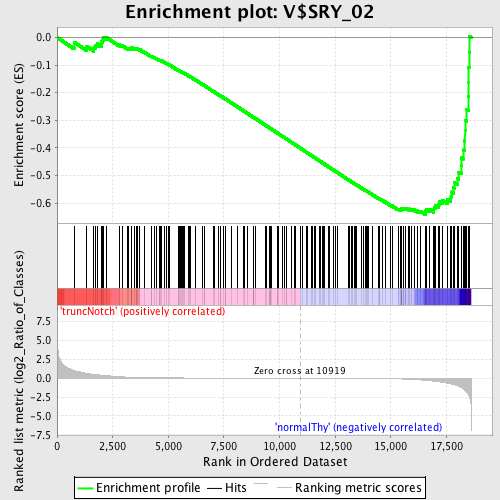
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | V$SRY_02 |
| Enrichment Score (ES) | -0.6409024 |
| Normalized Enrichment Score (NES) | -1.6852388 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00970693 |
| FWER p-Value | 0.062 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC12A8 | 4810092 | 763 | 1.021 | -0.0169 | No | ||
| 2 | E2F1 | 5360093 | 1330 | 0.638 | -0.0323 | No | ||
| 3 | PRKAG1 | 6450091 | 1656 | 0.503 | -0.0379 | No | ||
| 4 | AP4M1 | 70609 3440128 | 1716 | 0.481 | -0.0296 | No | ||
| 5 | OLFML3 | 7100463 | 1802 | 0.454 | -0.0233 | No | ||
| 6 | LMO4 | 3800746 | 2000 | 0.395 | -0.0245 | No | ||
| 7 | RBBP4 | 6650528 | 2010 | 0.392 | -0.0156 | No | ||
| 8 | LRP1 | 6270386 | 2060 | 0.378 | -0.0092 | No | ||
| 9 | CAMK1G | 1190037 6550315 | 2083 | 0.373 | -0.0015 | No | ||
| 10 | MYST3 | 5270500 | 2215 | 0.339 | -0.0004 | No | ||
| 11 | TGFB3 | 1070041 | 2790 | 0.197 | -0.0268 | No | ||
| 12 | DHX40 | 360672 | 2916 | 0.177 | -0.0294 | No | ||
| 13 | HESX1 | 4050746 | 3174 | 0.142 | -0.0399 | No | ||
| 14 | CCBL1 | 2350082 | 3223 | 0.137 | -0.0392 | No | ||
| 15 | MPP6 | 6020148 | 3324 | 0.125 | -0.0416 | No | ||
| 16 | FBN2 | 50435 | 3332 | 0.124 | -0.0391 | No | ||
| 17 | TNFRSF19 | 3870731 6940730 | 3361 | 0.120 | -0.0377 | No | ||
| 18 | MXI1 | 5050064 5130484 | 3466 | 0.111 | -0.0407 | No | ||
| 19 | SLC7A1 | 2450193 2710241 | 3547 | 0.105 | -0.0425 | No | ||
| 20 | TBX6 | 4730139 6840647 | 3562 | 0.103 | -0.0408 | No | ||
| 21 | DTNA | 1340600 1780731 2340278 2850132 | 3603 | 0.100 | -0.0406 | No | ||
| 22 | A2BP1 | 2370390 4590593 5550014 | 3723 | 0.091 | -0.0448 | No | ||
| 23 | MRC2 | 110369 2480520 | 3923 | 0.080 | -0.0537 | No | ||
| 24 | MCM7 | 3290292 5220056 | 4238 | 0.065 | -0.0692 | No | ||
| 25 | FOXP2 | 3520561 4150372 4760524 | 4262 | 0.064 | -0.0689 | No | ||
| 26 | POU3F4 | 870274 | 4364 | 0.059 | -0.0729 | No | ||
| 27 | SP8 | 4060576 | 4456 | 0.057 | -0.0765 | No | ||
| 28 | PTGFRN | 4120524 | 4599 | 0.052 | -0.0829 | No | ||
| 29 | SLC26A9 | 3440722 5340088 7510093 | 4611 | 0.052 | -0.0823 | No | ||
| 30 | SOX6 | 2940458 4070601 6840717 | 4638 | 0.051 | -0.0825 | No | ||
| 31 | HLX1 | 4250102 | 4704 | 0.049 | -0.0848 | No | ||
| 32 | NFIB | 460450 | 4827 | 0.045 | -0.0904 | No | ||
| 33 | NTF3 | 5890131 | 4933 | 0.043 | -0.0950 | No | ||
| 34 | RAPGEF5 | 1500603 3450427 6130056 | 4999 | 0.041 | -0.0976 | No | ||
| 35 | EMCN | 5130672 | 5037 | 0.041 | -0.0986 | No | ||
| 36 | TIAL1 | 4150048 6510605 | 5446 | 0.032 | -0.1199 | No | ||
| 37 | CLDN18 | 2030193 | 5456 | 0.032 | -0.1196 | No | ||
| 38 | FOXB1 | 4920270 5290463 | 5502 | 0.031 | -0.1213 | No | ||
| 39 | FGF9 | 1050195 | 5564 | 0.030 | -0.1239 | No | ||
| 40 | RHOBTB1 | 1410010 | 5568 | 0.030 | -0.1234 | No | ||
| 41 | LRP5 | 2100397 3170484 | 5622 | 0.029 | -0.1255 | No | ||
| 42 | OLFM1 | 3360161 5570066 | 5696 | 0.028 | -0.1288 | No | ||
| 43 | ANGPT1 | 3990368 5130288 6770035 | 5725 | 0.028 | -0.1297 | No | ||
| 44 | HOXC4 | 1940193 | 5909 | 0.025 | -0.1390 | No | ||
| 45 | EBF2 | 940324 | 5936 | 0.025 | -0.1398 | No | ||
| 46 | STC1 | 360161 | 5992 | 0.024 | -0.1422 | No | ||
| 47 | RBM8A | 630168 6220538 | 6232 | 0.021 | -0.1547 | No | ||
| 48 | PTN | 5910161 | 6525 | 0.018 | -0.1701 | No | ||
| 49 | NFIA | 2760129 5860278 | 6607 | 0.018 | -0.1740 | No | ||
| 50 | RBM9 | 2510092 6290088 | 6616 | 0.018 | -0.1740 | No | ||
| 51 | SOX5 | 2370576 2900167 3190128 5050528 | 7032 | 0.014 | -0.1962 | No | ||
| 52 | ELAVL2 | 360181 | 7089 | 0.014 | -0.1989 | No | ||
| 53 | NFKBIE | 580390 2190086 | 7265 | 0.013 | -0.2081 | No | ||
| 54 | PIK3R1 | 4730671 | 7321 | 0.013 | -0.2107 | No | ||
| 55 | SEMA6C | 460458 1580110 | 7322 | 0.013 | -0.2104 | No | ||
| 56 | MID2 | 4120309 | 7496 | 0.012 | -0.2195 | No | ||
| 57 | TBX5 | 2900132 6370484 | 7553 | 0.011 | -0.2223 | No | ||
| 58 | NKX2-2 | 4150731 | 7557 | 0.011 | -0.2222 | No | ||
| 59 | ESRRG | 4010181 | 7822 | 0.010 | -0.2363 | No | ||
| 60 | TECTA | 2570372 | 8114 | 0.009 | -0.2518 | No | ||
| 61 | SOX2 | 630487 2230491 | 8394 | 0.008 | -0.2668 | No | ||
| 62 | NR3C2 | 2480324 6400093 | 8408 | 0.008 | -0.2673 | No | ||
| 63 | HOXB5 | 2900538 | 8568 | 0.007 | -0.2757 | No | ||
| 64 | KERA | 6350136 | 8813 | 0.006 | -0.2888 | No | ||
| 65 | GATA2 | 6590280 | 8826 | 0.006 | -0.2893 | No | ||
| 66 | APBA1 | 3140487 | 8844 | 0.006 | -0.2901 | No | ||
| 67 | POU4F1 | 2260315 | 8938 | 0.006 | -0.2950 | No | ||
| 68 | LPHN2 | 4480010 | 9376 | 0.004 | -0.3186 | No | ||
| 69 | HOXD10 | 6100039 | 9394 | 0.004 | -0.3194 | No | ||
| 70 | LRRN1 | 3290154 | 9525 | 0.004 | -0.3264 | No | ||
| 71 | NR4A1 | 6290161 | 9539 | 0.004 | -0.3270 | No | ||
| 72 | SIX4 | 6760022 | 9583 | 0.004 | -0.3292 | No | ||
| 73 | DLL1 | 1770377 | 9609 | 0.004 | -0.3305 | No | ||
| 74 | LRCH2 | 2900162 | 9647 | 0.004 | -0.3324 | No | ||
| 75 | PURA | 3870156 | 9895 | 0.003 | -0.3457 | No | ||
| 76 | PAK3 | 4210136 | 9922 | 0.003 | -0.3471 | No | ||
| 77 | QKI | 5220093 6130044 | 9948 | 0.003 | -0.3483 | No | ||
| 78 | RCN1 | 2480041 | 10117 | 0.002 | -0.3574 | No | ||
| 79 | GPR110 | 1190215 | 10229 | 0.002 | -0.3634 | No | ||
| 80 | ZIC2 | 1410408 | 10312 | 0.002 | -0.3678 | No | ||
| 81 | MAB21L1 | 1450634 1580092 | 10316 | 0.002 | -0.3679 | No | ||
| 82 | EPHA7 | 1190288 4010278 | 10549 | 0.001 | -0.3804 | No | ||
| 83 | FST | 1110600 | 10668 | 0.001 | -0.3868 | No | ||
| 84 | BNC2 | 4810603 | 10701 | 0.001 | -0.3885 | No | ||
| 85 | MOBP | 7000059 | 10928 | -0.000 | -0.4008 | No | ||
| 86 | CD36 | 5890575 | 11024 | -0.000 | -0.4059 | No | ||
| 87 | MLLT7 | 4480707 | 11196 | -0.001 | -0.4152 | No | ||
| 88 | B3GALT2 | 430136 | 11264 | -0.001 | -0.4188 | No | ||
| 89 | FGFR2 | 4670601 5570402 6940377 | 11419 | -0.001 | -0.4271 | No | ||
| 90 | KLHL13 | 6590731 | 11471 | -0.002 | -0.4298 | No | ||
| 91 | TCF15 | 1690400 | 11590 | -0.002 | -0.4362 | No | ||
| 92 | IL5RA | 4540091 | 11633 | -0.002 | -0.4384 | No | ||
| 93 | STAG2 | 4540132 | 11810 | -0.003 | -0.4479 | No | ||
| 94 | H2AFY | 4730750 | 11833 | -0.003 | -0.4490 | No | ||
| 95 | BAMBI | 6650463 | 11938 | -0.003 | -0.4546 | No | ||
| 96 | NUDT11 | 3840239 | 11986 | -0.003 | -0.4570 | No | ||
| 97 | DEPDC2 | 5220102 | 12023 | -0.004 | -0.4589 | No | ||
| 98 | EN2 | 7000368 | 12196 | -0.004 | -0.4681 | No | ||
| 99 | ABCA6 | 130538 1050706 | 12234 | -0.004 | -0.4700 | No | ||
| 100 | CRH | 3710301 | 12429 | -0.005 | -0.4804 | No | ||
| 101 | CCNJ | 870047 | 12505 | -0.006 | -0.4843 | No | ||
| 102 | TCF1 | 5390022 | 12587 | -0.006 | -0.4886 | No | ||
| 103 | GNAQ | 430670 4210131 5900736 | 13080 | -0.009 | -0.5150 | No | ||
| 104 | SLC13A1 | 5220736 | 13126 | -0.009 | -0.5173 | No | ||
| 105 | MITF | 380056 | 13233 | -0.010 | -0.5228 | No | ||
| 106 | CHN1 | 6040441 | 13272 | -0.010 | -0.5246 | No | ||
| 107 | GRAP | 1770195 | 13367 | -0.011 | -0.5294 | No | ||
| 108 | ALDH1A2 | 2320301 | 13414 | -0.011 | -0.5317 | No | ||
| 109 | MDGA1 | 1170139 | 13439 | -0.012 | -0.5327 | No | ||
| 110 | HS3ST1 | 1850193 | 13693 | -0.014 | -0.5460 | No | ||
| 111 | LEMD1 | 3990132 5720187 | 13759 | -0.015 | -0.5492 | No | ||
| 112 | PAK1 | 4540315 | 13867 | -0.017 | -0.5546 | No | ||
| 113 | DHRS3 | 360609 | 13924 | -0.018 | -0.5572 | No | ||
| 114 | GNAO1 | 2510292 6590487 | 13937 | -0.018 | -0.5574 | No | ||
| 115 | MYLK | 4010600 7000364 | 13974 | -0.019 | -0.5589 | No | ||
| 116 | CACNA1E | 840253 | 14161 | -0.022 | -0.5685 | No | ||
| 117 | NR4A3 | 2900021 5860095 5910039 | 14428 | -0.029 | -0.5822 | No | ||
| 118 | PCF11 | 1240180 | 14480 | -0.031 | -0.5843 | No | ||
| 119 | MLLT3 | 510204 3850722 6200088 | 14508 | -0.032 | -0.5850 | No | ||
| 120 | HEY1 | 4570546 | 14635 | -0.037 | -0.5909 | No | ||
| 121 | CFL2 | 380239 6200368 | 14738 | -0.043 | -0.5954 | No | ||
| 122 | FN1 | 1170601 2970647 6220288 6940037 | 14972 | -0.055 | -0.6067 | No | ||
| 123 | ELAVL4 | 50735 3360086 5220167 | 15058 | -0.061 | -0.6098 | No | ||
| 124 | ARHGAP15 | 610446 2340327 2900438 | 15324 | -0.083 | -0.6222 | No | ||
| 125 | KCNN2 | 990025 3520524 | 15423 | -0.093 | -0.6253 | No | ||
| 126 | TNFRSF19L | 1170168 1190358 5670270 | 15459 | -0.096 | -0.6249 | No | ||
| 127 | COMMD3 | 2510259 | 15462 | -0.096 | -0.6227 | No | ||
| 128 | TCF4 | 520021 | 15471 | -0.097 | -0.6208 | No | ||
| 129 | CPNE1 | 3440131 | 15497 | -0.101 | -0.6198 | No | ||
| 130 | MBNL2 | 3450707 | 15577 | -0.109 | -0.6214 | No | ||
| 131 | MSI2 | 1190066 5270039 | 15578 | -0.109 | -0.6188 | No | ||
| 132 | PHF16 | 50373 7200161 | 15653 | -0.117 | -0.6200 | No | ||
| 133 | ZADH2 | 520438 | 15678 | -0.120 | -0.6185 | No | ||
| 134 | TACSTD2 | 6660341 | 15798 | -0.134 | -0.6217 | No | ||
| 135 | AP1G2 | 780458 2690068 | 15846 | -0.140 | -0.6209 | No | ||
| 136 | ICK | 1580746 3140021 | 15946 | -0.156 | -0.6225 | No | ||
| 137 | NANOS1 | 1300402 | 16056 | -0.171 | -0.6244 | No | ||
| 138 | ZFYVE1 | 60440 | 16215 | -0.199 | -0.6282 | No | ||
| 139 | PELI2 | 1940364 | 16317 | -0.219 | -0.6284 | No | ||
| 140 | NSF | 4070075 | 16549 | -0.278 | -0.6343 | Yes | ||
| 141 | PUM1 | 6130500 | 16560 | -0.279 | -0.6281 | Yes | ||
| 142 | PIAS1 | 5340301 | 16608 | -0.296 | -0.6236 | Yes | ||
| 143 | MYO18A | 1230333 | 16755 | -0.336 | -0.6234 | Yes | ||
| 144 | YWHAG | 3780341 | 16937 | -0.391 | -0.6239 | Yes | ||
| 145 | SMAD3 | 6450671 | 16940 | -0.392 | -0.6146 | Yes | ||
| 146 | CHD1 | 4590048 | 17013 | -0.419 | -0.6085 | Yes | ||
| 147 | NFIX | 2450152 | 17139 | -0.468 | -0.6040 | Yes | ||
| 148 | CTLA4 | 6590537 | 17187 | -0.486 | -0.5950 | Yes | ||
| 149 | LDB1 | 5270601 | 17329 | -0.545 | -0.5895 | Yes | ||
| 150 | ULK1 | 6100315 | 17557 | -0.663 | -0.5860 | Yes | ||
| 151 | DDX6 | 2630025 6770408 | 17703 | -0.744 | -0.5760 | Yes | ||
| 152 | TIAM1 | 5420288 | 17739 | -0.768 | -0.5595 | Yes | ||
| 153 | WASL | 3290170 | 17820 | -0.826 | -0.5441 | Yes | ||
| 154 | PIK3CG | 5890110 | 17858 | -0.857 | -0.5256 | Yes | ||
| 155 | EIF4G2 | 3800575 6860184 | 17993 | -0.974 | -0.5095 | Yes | ||
| 156 | SFRS7 | 2760408 | 18057 | -1.040 | -0.4880 | Yes | ||
| 157 | XKRX | 3800192 | 18168 | -1.208 | -0.4650 | Yes | ||
| 158 | NFE2L1 | 6130053 | 18175 | -1.214 | -0.4362 | Yes | ||
| 159 | PPM1D | 1690193 | 18249 | -1.366 | -0.4075 | Yes | ||
| 160 | KLF7 | 50390 | 18299 | -1.477 | -0.3748 | Yes | ||
| 161 | KBTBD2 | 2320014 | 18334 | -1.590 | -0.3385 | Yes | ||
| 162 | CDC42EP3 | 2480138 | 18338 | -1.617 | -0.3000 | Yes | ||
| 163 | MAML1 | 2760008 | 18415 | -1.816 | -0.2606 | Yes | ||
| 164 | HSD11B1 | 450066 5550408 | 18477 | -2.066 | -0.2144 | Yes | ||
| 165 | CSRP2BP | 4570717 5550333 | 18495 | -2.214 | -0.1623 | Yes | ||
| 166 | DUSP6 | 5910286 7100070 | 18498 | -2.236 | -0.1089 | Yes | ||
| 167 | SATB1 | 5670154 | 18523 | -2.380 | -0.0532 | Yes | ||
| 168 | KLF3 | 5130438 | 18530 | -2.428 | 0.0047 | Yes |