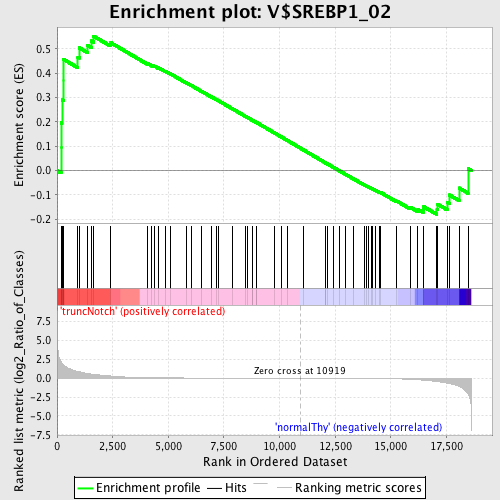
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$SREBP1_02 |
| Enrichment Score (ES) | 0.55280054 |
| Normalized Enrichment Score (NES) | 1.2049267 |
| Nominal p-value | 0.15050167 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DPYSL2 | 2100427 3130112 5700324 | 180 | 2.110 | 0.0941 | Yes | ||
| 2 | CEBPB | 2970019 | 183 | 2.087 | 0.1967 | Yes | ||
| 3 | NDRG2 | 450403 | 220 | 1.946 | 0.2905 | Yes | ||
| 4 | RAB33A | 6590195 | 284 | 1.732 | 0.3723 | Yes | ||
| 5 | POU2AF1 | 5690242 | 285 | 1.731 | 0.4574 | Yes | ||
| 6 | SREBF2 | 3390692 | 933 | 0.890 | 0.4664 | Yes | ||
| 7 | GNAS | 630441 1850373 4050152 | 1003 | 0.849 | 0.5044 | Yes | ||
| 8 | HOXB1 | 2120110 | 1373 | 0.618 | 0.5150 | Yes | ||
| 9 | LOXL1 | 3520537 | 1537 | 0.547 | 0.5331 | Yes | ||
| 10 | SPI1 | 1410397 | 1639 | 0.511 | 0.5528 | Yes | ||
| 11 | CACNA2D3 | 4010184 | 2386 | 0.295 | 0.5271 | No | ||
| 12 | PACSIN3 | 4540369 4540400 5360037 6940129 | 4061 | 0.073 | 0.4405 | No | ||
| 13 | FOXP2 | 3520561 4150372 4760524 | 4262 | 0.064 | 0.4329 | No | ||
| 14 | KIF1C | 2480484 | 4379 | 0.059 | 0.4295 | No | ||
| 15 | DDR1 | 2060044 5220180 | 4542 | 0.054 | 0.4235 | No | ||
| 16 | GNB2 | 2350053 | 4878 | 0.044 | 0.4076 | No | ||
| 17 | HOXB8 | 3710450 | 5077 | 0.040 | 0.3989 | No | ||
| 18 | RHOG | 6760575 | 5798 | 0.026 | 0.3614 | No | ||
| 19 | SSTR3 | 5420064 | 6027 | 0.023 | 0.3502 | No | ||
| 20 | CD2AP | 1940369 | 6506 | 0.019 | 0.3254 | No | ||
| 21 | PHOX2B | 5270075 | 6935 | 0.015 | 0.3031 | No | ||
| 22 | BCL6B | 60047 | 7147 | 0.014 | 0.2924 | No | ||
| 23 | GFAP | 2060092 | 7266 | 0.013 | 0.2866 | No | ||
| 24 | CPA4 | 3610100 | 7873 | 0.010 | 0.2545 | No | ||
| 25 | DLGAP4 | 4570672 5690161 | 8446 | 0.007 | 0.2240 | No | ||
| 26 | BAI3 | 940692 | 8567 | 0.007 | 0.2179 | No | ||
| 27 | NOL4 | 5050446 5360519 | 8769 | 0.006 | 0.2074 | No | ||
| 28 | SEZ6 | 3450722 | 8801 | 0.006 | 0.2060 | No | ||
| 29 | INPP4A | 510215 1090541 4850093 6100519 | 8956 | 0.006 | 0.1980 | No | ||
| 30 | POU4F2 | 2120195 2570022 | 8983 | 0.006 | 0.1968 | No | ||
| 31 | RBP3 | 1400059 | 9778 | 0.003 | 0.1542 | No | ||
| 32 | HYAL1 | 3850341 | 10088 | 0.002 | 0.1377 | No | ||
| 33 | MEF2C | 670025 780338 | 10376 | 0.002 | 0.1223 | No | ||
| 34 | NTN4 | 6940398 | 11075 | -0.000 | 0.0847 | No | ||
| 35 | GREM1 | 3940180 | 12073 | -0.004 | 0.0312 | No | ||
| 36 | PLA2G3 | 5130739 | 12080 | -0.004 | 0.0310 | No | ||
| 37 | FGF12 | 1740446 2360037 | 12167 | -0.004 | 0.0266 | No | ||
| 38 | ATP2B2 | 3780397 | 12439 | -0.005 | 0.0122 | No | ||
| 39 | TOM1L1 | 4920056 3190070 | 12701 | -0.006 | -0.0015 | No | ||
| 40 | NF1 | 6980433 | 12948 | -0.008 | -0.0144 | No | ||
| 41 | HS6ST2 | 520133 5550603 | 13322 | -0.011 | -0.0340 | No | ||
| 42 | PLAG1 | 1450142 3870139 6520039 | 13805 | -0.016 | -0.0592 | No | ||
| 43 | DHRS3 | 360609 | 13924 | -0.018 | -0.0646 | No | ||
| 44 | WDR13 | 3840520 4670064 | 14007 | -0.019 | -0.0681 | No | ||
| 45 | CHRM3 | 6110725 | 14116 | -0.021 | -0.0729 | No | ||
| 46 | UBE2B | 4780497 | 14177 | -0.022 | -0.0751 | No | ||
| 47 | NOTCH2 | 2570397 | 14297 | -0.025 | -0.0803 | No | ||
| 48 | YWHAE | 5310435 | 14503 | -0.032 | -0.0897 | No | ||
| 49 | NRGN | 4280433 | 14541 | -0.033 | -0.0901 | No | ||
| 50 | KTN1 | 70446 3450609 4560048 | 15261 | -0.076 | -0.1251 | No | ||
| 51 | EIF5A | 1500059 5290022 6370026 | 15867 | -0.144 | -0.1506 | No | ||
| 52 | ZFYVE1 | 60440 | 16215 | -0.199 | -0.1596 | No | ||
| 53 | ELOVL5 | 3800170 | 16452 | -0.250 | -0.1600 | No | ||
| 54 | RARA | 4050161 | 16482 | -0.260 | -0.1487 | No | ||
| 55 | MORF4L2 | 6450133 | 17074 | -0.438 | -0.1590 | No | ||
| 56 | BAZ2A | 730184 | 17111 | -0.455 | -0.1386 | No | ||
| 57 | CORO1A | 3140609 3190020 3190037 | 17532 | -0.649 | -0.1293 | No | ||
| 58 | PTMA | 5570148 | 17633 | -0.699 | -0.1003 | No | ||
| 59 | FBS1 | 2570520 | 18070 | -1.061 | -0.0716 | No | ||
| 60 | ZFP36L1 | 2510138 4120048 | 18473 | -2.052 | 0.0077 | No |

