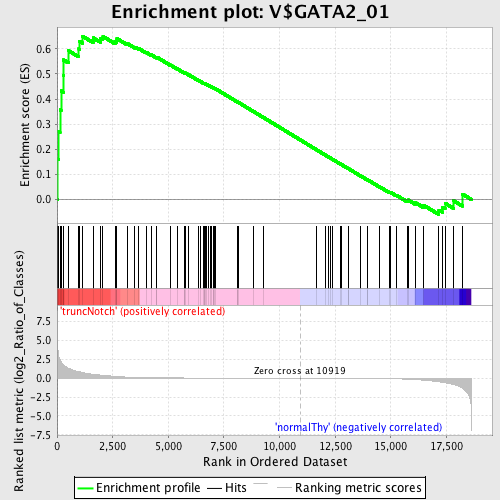
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$GATA2_01 |
| Enrichment Score (ES) | 0.65052587 |
| Normalized Enrichment Score (NES) | 1.4467473 |
| Nominal p-value | 0.0208 |
| FDR q-value | 0.6401038 |
| FWER p-Value | 0.985 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SFRS2 | 50707 380593 | 24 | 4.382 | 0.1610 | Yes | ||
| 2 | PDLIM4 | 3990358 | 67 | 3.030 | 0.2710 | Yes | ||
| 3 | UBTF | 4210450 6840403 | 132 | 2.409 | 0.3568 | Yes | ||
| 4 | DPYSL2 | 2100427 3130112 5700324 | 180 | 2.110 | 0.4324 | Yes | ||
| 5 | UBE2H | 1980142 2970079 | 277 | 1.757 | 0.4923 | Yes | ||
| 6 | NCDN | 50040 1980739 3520603 | 287 | 1.726 | 0.5557 | Yes | ||
| 7 | ABCB6 | 6350138 | 509 | 1.319 | 0.5926 | Yes | ||
| 8 | RAB34 | 770411 | 978 | 0.863 | 0.5994 | Yes | ||
| 9 | GNAS | 630441 1850373 4050152 | 1003 | 0.849 | 0.6295 | Yes | ||
| 10 | PTK7 | 380315 | 1135 | 0.757 | 0.6505 | Yes | ||
| 11 | PPOX | 2640678 3850102 | 1621 | 0.518 | 0.6435 | No | ||
| 12 | FKBP2 | 2320025 | 1944 | 0.417 | 0.6416 | No | ||
| 13 | LRP1 | 6270386 | 2060 | 0.378 | 0.6494 | No | ||
| 14 | NEURL | 5390538 | 2611 | 0.237 | 0.6285 | No | ||
| 15 | OAZ2 | 2810110 | 2682 | 0.221 | 0.6330 | No | ||
| 16 | PPP2R2A | 2900014 5700575 | 2688 | 0.220 | 0.6408 | No | ||
| 17 | ABHD8 | 6660082 | 3161 | 0.144 | 0.6207 | No | ||
| 18 | POLD4 | 2320082 | 3491 | 0.109 | 0.6070 | No | ||
| 19 | CNNM1 | 2190433 | 3658 | 0.096 | 0.6016 | No | ||
| 20 | FBXW7 | 4210338 7050280 | 4001 | 0.076 | 0.5860 | No | ||
| 21 | UBXD3 | 5050193 | 4220 | 0.066 | 0.5767 | No | ||
| 22 | PHF6 | 3120632 3990041 | 4473 | 0.056 | 0.5652 | No | ||
| 23 | HOXA10 | 6110397 7100458 | 4487 | 0.056 | 0.5665 | No | ||
| 24 | DPF3 | 2650128 2970168 | 5088 | 0.039 | 0.5357 | No | ||
| 25 | NXPH3 | 5860292 | 5395 | 0.033 | 0.5204 | No | ||
| 26 | ANGPT1 | 3990368 5130288 6770035 | 5725 | 0.028 | 0.5037 | No | ||
| 27 | PDGFRA | 2940332 | 5734 | 0.028 | 0.5043 | No | ||
| 28 | AMOT | 1940121 2940647 6590093 | 5762 | 0.027 | 0.5038 | No | ||
| 29 | WNT5A | 840685 3120152 | 5922 | 0.025 | 0.4962 | No | ||
| 30 | SYNCRIP | 1690195 3140113 4670279 | 6344 | 0.020 | 0.4742 | No | ||
| 31 | GRIK2 | 4610164 | 6454 | 0.019 | 0.4690 | No | ||
| 32 | CNOT3 | 130519 4480292 1990609 | 6569 | 0.018 | 0.4635 | No | ||
| 33 | NFIA | 2760129 5860278 | 6607 | 0.018 | 0.4622 | No | ||
| 34 | SYN1 | 4920301 7050440 | 6630 | 0.017 | 0.4617 | No | ||
| 35 | GRIN2A | 6550538 | 6663 | 0.017 | 0.4606 | No | ||
| 36 | SALL1 | 5420020 7050195 | 6705 | 0.017 | 0.4590 | No | ||
| 37 | REPS2 | 2690066 | 6809 | 0.016 | 0.4540 | No | ||
| 38 | SLC39A5 | 2900100 4610440 | 6891 | 0.016 | 0.4502 | No | ||
| 39 | PHOX2B | 5270075 | 6935 | 0.015 | 0.4485 | No | ||
| 40 | HNRPL | 4210593 | 7018 | 0.015 | 0.4446 | No | ||
| 41 | RAB3C | 5420072 | 7024 | 0.014 | 0.4449 | No | ||
| 42 | VASP | 7050500 | 7056 | 0.014 | 0.4437 | No | ||
| 43 | HOXB6 | 4210739 | 7129 | 0.014 | 0.4403 | No | ||
| 44 | PLXNC1 | 5860315 | 8113 | 0.009 | 0.3877 | No | ||
| 45 | PAFAH1B1 | 4230333 6420121 6450066 | 8128 | 0.009 | 0.3872 | No | ||
| 46 | ST7L | 840735 2360576 | 8142 | 0.009 | 0.3868 | No | ||
| 47 | LHX6 | 2360731 3360397 | 8832 | 0.006 | 0.3499 | No | ||
| 48 | GRID2 | 2680242 | 9256 | 0.005 | 0.3273 | No | ||
| 49 | RBMS3 | 6110722 | 11650 | -0.002 | 0.1983 | No | ||
| 50 | GREM1 | 3940180 | 12073 | -0.004 | 0.1757 | No | ||
| 51 | HS6ST3 | 5910138 | 12186 | -0.004 | 0.1698 | No | ||
| 52 | SP3 | 3840338 | 12266 | -0.004 | 0.1657 | No | ||
| 53 | POU3F1 | 3710022 | 12375 | -0.005 | 0.1601 | No | ||
| 54 | SP7 | 1050315 | 12738 | -0.007 | 0.1408 | No | ||
| 55 | SLC8A3 | 4120711 6290592 | 12774 | -0.007 | 0.1392 | No | ||
| 56 | FOXA3 | 2680121 | 13091 | -0.009 | 0.1224 | No | ||
| 57 | FBXW11 | 6450632 | 13100 | -0.009 | 0.1223 | No | ||
| 58 | CAPZA1 | 3140390 | 13643 | -0.014 | 0.0936 | No | ||
| 59 | FOXA2 | 540338 5860441 | 13945 | -0.018 | 0.0781 | No | ||
| 60 | MCTS1 | 6130133 | 13965 | -0.018 | 0.0777 | No | ||
| 61 | YWHAE | 5310435 | 14503 | -0.032 | 0.0499 | No | ||
| 62 | HDAC3 | 4060072 | 14956 | -0.055 | 0.0276 | No | ||
| 63 | BLCAP | 3990253 | 14992 | -0.057 | 0.0278 | No | ||
| 64 | RPL8 | 20296 | 15246 | -0.075 | 0.0169 | No | ||
| 65 | KLHL5 | 3060484 6400292 | 15750 | -0.129 | -0.0054 | No | ||
| 66 | CRMP1 | 4810014 | 15780 | -0.132 | -0.0021 | No | ||
| 67 | EIF4G1 | 4070446 | 16118 | -0.180 | -0.0136 | No | ||
| 68 | SRRM1 | 6110025 | 16479 | -0.258 | -0.0234 | No | ||
| 69 | KPNB1 | 1690138 | 17159 | -0.476 | -0.0424 | No | ||
| 70 | MPV17 | 1190133 4070427 4570577 | 17317 | -0.539 | -0.0309 | No | ||
| 71 | DNAJB5 | 2900215 | 17440 | -0.600 | -0.0153 | No | ||
| 72 | GATA3 | 6130068 | 17815 | -0.824 | -0.0049 | No | ||
| 73 | SPOP | 450035 | 18217 | -1.298 | 0.0215 | No |

