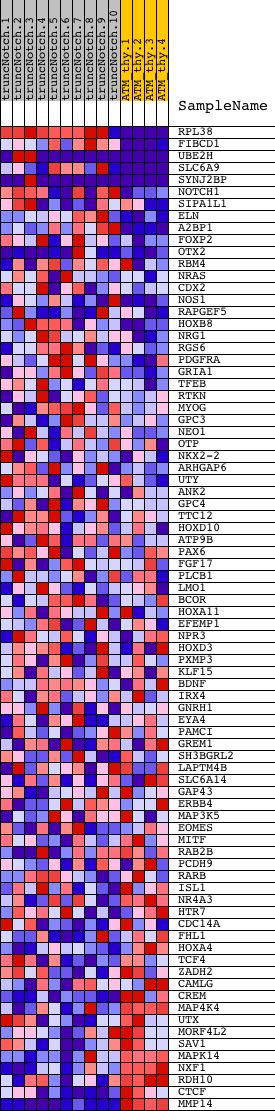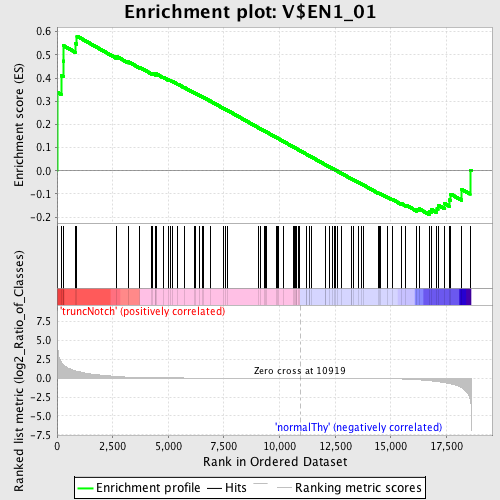
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$EN1_01 |
| Enrichment Score (ES) | 0.58060646 |
| Normalized Enrichment Score (NES) | 1.3031027 |
| Nominal p-value | 0.0625 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL38 | 70040 4060138 6200601 | 1 | 8.746 | 0.3389 | Yes | ||
| 2 | FIBCD1 | 1500500 | 182 | 2.094 | 0.4104 | Yes | ||
| 3 | UBE2H | 1980142 2970079 | 277 | 1.757 | 0.4734 | Yes | ||
| 4 | SLC6A9 | 2470520 | 292 | 1.714 | 0.5391 | Yes | ||
| 5 | SYNJ2BP | 4150672 6770706 | 813 | 0.978 | 0.5490 | Yes | ||
| 6 | NOTCH1 | 3390114 | 891 | 0.923 | 0.5806 | Yes | ||
| 7 | SIPA1L1 | 50292 | 2681 | 0.222 | 0.4927 | No | ||
| 8 | ELN | 5080347 | 3201 | 0.139 | 0.4701 | No | ||
| 9 | A2BP1 | 2370390 4590593 5550014 | 3723 | 0.091 | 0.4455 | No | ||
| 10 | FOXP2 | 3520561 4150372 4760524 | 4262 | 0.064 | 0.4190 | No | ||
| 11 | OTX2 | 1190471 | 4293 | 0.063 | 0.4198 | No | ||
| 12 | RBM4 | 2360113 5080750 | 4428 | 0.057 | 0.4148 | No | ||
| 13 | NRAS | 6900577 | 4430 | 0.057 | 0.4169 | No | ||
| 14 | CDX2 | 3850349 | 4454 | 0.057 | 0.4179 | No | ||
| 15 | NOS1 | 5860129 | 4766 | 0.047 | 0.4029 | No | ||
| 16 | RAPGEF5 | 1500603 3450427 6130056 | 4999 | 0.041 | 0.3920 | No | ||
| 17 | HOXB8 | 3710450 | 5077 | 0.040 | 0.3894 | No | ||
| 18 | NRG1 | 1050332 | 5184 | 0.037 | 0.3851 | No | ||
| 19 | RGS6 | 6040601 | 5425 | 0.033 | 0.3735 | No | ||
| 20 | PDGFRA | 2940332 | 5734 | 0.028 | 0.3579 | No | ||
| 21 | GRIA1 | 1340152 3780750 4920440 | 6155 | 0.022 | 0.3361 | No | ||
| 22 | TFEB | 3390121 | 6226 | 0.021 | 0.3331 | No | ||
| 23 | RTKN | 5910253 6380082 | 6400 | 0.019 | 0.3246 | No | ||
| 24 | MYOG | 3190672 | 6547 | 0.018 | 0.3174 | No | ||
| 25 | GPC3 | 6180735 | 6598 | 0.018 | 0.3154 | No | ||
| 26 | NEO1 | 1990154 | 6875 | 0.016 | 0.3011 | No | ||
| 27 | OTP | 2630070 | 7497 | 0.012 | 0.2680 | No | ||
| 28 | NKX2-2 | 4150731 | 7557 | 0.011 | 0.2653 | No | ||
| 29 | ARHGAP6 | 2060121 | 7659 | 0.011 | 0.2603 | No | ||
| 30 | UTY | 5890441 | 9072 | 0.005 | 0.1843 | No | ||
| 31 | ANK2 | 6510546 | 9151 | 0.005 | 0.1803 | No | ||
| 32 | GPC4 | 5910408 | 9341 | 0.004 | 0.1703 | No | ||
| 33 | TTC12 | 4480632 | 9353 | 0.004 | 0.1698 | No | ||
| 34 | HOXD10 | 6100039 | 9394 | 0.004 | 0.1678 | No | ||
| 35 | ATP9B | 3710494 6020239 | 9874 | 0.003 | 0.1421 | No | ||
| 36 | PAX6 | 1190025 | 9898 | 0.003 | 0.1410 | No | ||
| 37 | FGF17 | 3130022 | 9959 | 0.003 | 0.1379 | No | ||
| 38 | PLCB1 | 2570050 7100102 | 10158 | 0.002 | 0.1273 | No | ||
| 39 | LMO1 | 3170528 | 10605 | 0.001 | 0.1032 | No | ||
| 40 | BCOR | 1660452 3940053 | 10650 | 0.001 | 0.1009 | No | ||
| 41 | HOXA11 | 6980133 | 10713 | 0.001 | 0.0976 | No | ||
| 42 | EFEMP1 | 5860324 | 10742 | 0.001 | 0.0961 | No | ||
| 43 | NPR3 | 1580239 | 10768 | 0.000 | 0.0947 | No | ||
| 44 | HOXD3 | 6450154 | 10852 | 0.000 | 0.0903 | No | ||
| 45 | PXMP3 | 2100088 1400458 6650671 | 10875 | 0.000 | 0.0891 | No | ||
| 46 | KLF15 | 4230164 | 11199 | -0.001 | 0.0717 | No | ||
| 47 | BDNF | 2940128 3520368 | 11224 | -0.001 | 0.0704 | No | ||
| 48 | IRX4 | 1980102 | 11344 | -0.001 | 0.0641 | No | ||
| 49 | GNRH1 | 430167 | 11347 | -0.001 | 0.0640 | No | ||
| 50 | EYA4 | 5360286 | 11418 | -0.001 | 0.0603 | No | ||
| 51 | PAMCI | 2320687 | 12060 | -0.004 | 0.0258 | No | ||
| 52 | GREM1 | 3940180 | 12073 | -0.004 | 0.0253 | No | ||
| 53 | SH3BGRL2 | 1780239 | 12255 | -0.004 | 0.0157 | No | ||
| 54 | LAPTM4B | 2060377 4150398 | 12399 | -0.005 | 0.0082 | No | ||
| 55 | SLC6A14 | 380368 | 12447 | -0.005 | 0.0059 | No | ||
| 56 | GAP43 | 3130504 | 12528 | -0.006 | 0.0018 | No | ||
| 57 | ERBB4 | 4610400 4810017 | 12582 | -0.006 | -0.0008 | No | ||
| 58 | MAP3K5 | 6020041 6380162 | 12783 | -0.007 | -0.0114 | No | ||
| 59 | EOMES | 2100707 | 13226 | -0.010 | -0.0348 | No | ||
| 60 | MITF | 380056 | 13233 | -0.010 | -0.0348 | No | ||
| 61 | RAB2B | 6200091 | 13308 | -0.010 | -0.0384 | No | ||
| 62 | PCDH9 | 2100136 | 13555 | -0.013 | -0.0511 | No | ||
| 63 | RARB | 430139 1410138 | 13691 | -0.014 | -0.0579 | No | ||
| 64 | ISL1 | 4810239 | 13779 | -0.016 | -0.0620 | No | ||
| 65 | NR4A3 | 2900021 5860095 5910039 | 14428 | -0.029 | -0.0958 | No | ||
| 66 | HTR7 | 2680176 | 14490 | -0.031 | -0.0979 | No | ||
| 67 | CDC14A | 4050132 | 14547 | -0.033 | -0.0996 | No | ||
| 68 | FHL1 | 60278 | 14840 | -0.049 | -0.1135 | No | ||
| 69 | HOXA4 | 940152 | 15064 | -0.062 | -0.1231 | No | ||
| 70 | TCF4 | 520021 | 15471 | -0.097 | -0.1412 | No | ||
| 71 | ZADH2 | 520438 | 15678 | -0.120 | -0.1477 | No | ||
| 72 | CAMLG | 1410184 | 16174 | -0.191 | -0.1670 | No | ||
| 73 | CREM | 840156 6380438 6660041 6660168 | 16266 | -0.211 | -0.1638 | No | ||
| 74 | MAP4K4 | 2360059 | 16726 | -0.329 | -0.1758 | No | ||
| 75 | UTX | 2900017 6770452 | 16831 | -0.357 | -0.1675 | No | ||
| 76 | MORF4L2 | 6450133 | 17074 | -0.438 | -0.1636 | No | ||
| 77 | SAV1 | 5420541 | 17122 | -0.462 | -0.1482 | No | ||
| 78 | MAPK14 | 5290731 | 17410 | -0.583 | -0.1412 | No | ||
| 79 | NXF1 | 1570377 | 17627 | -0.697 | -0.1258 | No | ||
| 80 | RDH10 | 6650070 | 17692 | -0.737 | -0.1007 | No | ||
| 81 | CTCF | 5340017 | 18177 | -1.215 | -0.0797 | No | ||
| 82 | MMP14 | 670079 | 18560 | -2.666 | 0.0030 | No |