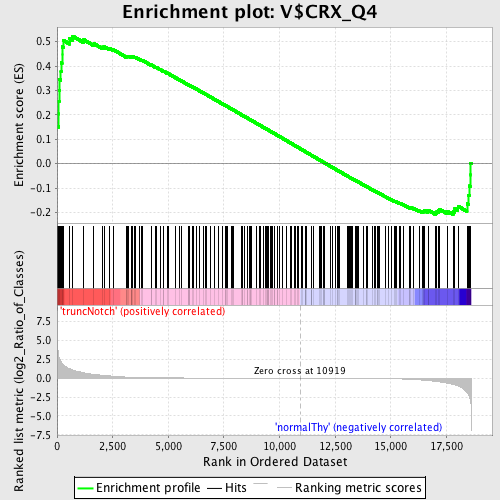
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | V$CRX_Q4 |
| Enrichment Score (ES) | 0.52244544 |
| Normalized Enrichment Score (NES) | 1.2728026 |
| Nominal p-value | 0.03748126 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TKT | 6590064 6860039 | 0 | 8.890 | 0.1530 | Yes | ||
| 2 | TCF7 | 3800736 5390181 | 60 | 3.131 | 0.2036 | Yes | ||
| 3 | TNNT1 | 3360671 | 66 | 3.049 | 0.2558 | Yes | ||
| 4 | ACAA2 | 1570347 2360324 6130139 | 91 | 2.687 | 0.3008 | Yes | ||
| 5 | CDKN2C | 5050750 5130148 | 102 | 2.582 | 0.3447 | Yes | ||
| 6 | HIST1H4C | 5570528 | 172 | 2.178 | 0.3784 | Yes | ||
| 7 | FIBCD1 | 1500500 | 182 | 2.094 | 0.4139 | Yes | ||
| 8 | ETV5 | 110017 | 222 | 1.935 | 0.4451 | Yes | ||
| 9 | DAP | 5270056 | 223 | 1.935 | 0.4784 | Yes | ||
| 10 | RAB33A | 6590195 | 284 | 1.732 | 0.5050 | Yes | ||
| 11 | MSH5 | 1340019 4730333 | 562 | 1.254 | 0.5115 | Yes | ||
| 12 | ANXA11 | 6370128 | 699 | 1.064 | 0.5224 | Yes | ||
| 13 | ESM1 | 1410594 | 1200 | 0.718 | 0.5077 | No | ||
| 14 | PRKAG1 | 6450091 | 1656 | 0.503 | 0.4916 | No | ||
| 15 | HMGN2 | 3140091 | 2032 | 0.386 | 0.4779 | No | ||
| 16 | BMI1 | 3830026 2630204 | 2123 | 0.361 | 0.4793 | No | ||
| 17 | TRIP10 | 460438 | 2368 | 0.299 | 0.4712 | No | ||
| 18 | ACADVL | 6040403 | 2513 | 0.261 | 0.4679 | No | ||
| 19 | WNT6 | 4570364 | 3099 | 0.152 | 0.4388 | No | ||
| 20 | NR2E1 | 4730288 | 3140 | 0.147 | 0.4391 | No | ||
| 21 | KCNA1 | 2030594 | 3165 | 0.143 | 0.4403 | No | ||
| 22 | RUNX3 | 3800546 | 3198 | 0.140 | 0.4410 | No | ||
| 23 | MERTK | 6420056 | 3326 | 0.125 | 0.4362 | No | ||
| 24 | RPS6KA3 | 1980707 | 3335 | 0.123 | 0.4379 | No | ||
| 25 | POLR2A | 5220647 | 3362 | 0.120 | 0.4386 | No | ||
| 26 | NR2F1 | 4120528 360402 | 3371 | 0.120 | 0.4402 | No | ||
| 27 | POLD4 | 2320082 | 3491 | 0.109 | 0.4356 | No | ||
| 28 | ZFP36L2 | 510180 | 3517 | 0.107 | 0.4361 | No | ||
| 29 | MIP | 6520333 | 3704 | 0.092 | 0.4276 | No | ||
| 30 | HOXC6 | 3850133 4920736 | 3792 | 0.088 | 0.4244 | No | ||
| 31 | PGF | 4810593 | 3817 | 0.086 | 0.4246 | No | ||
| 32 | UBXD3 | 5050193 | 4220 | 0.066 | 0.4039 | No | ||
| 33 | DLC1 | 1090632 6450594 | 4227 | 0.066 | 0.4047 | No | ||
| 34 | LRRTM4 | 5910100 | 4415 | 0.058 | 0.3956 | No | ||
| 35 | ZBTB4 | 6450441 | 4449 | 0.057 | 0.3947 | No | ||
| 36 | BDKRB1 | 730592 | 4639 | 0.051 | 0.3854 | No | ||
| 37 | NOS1 | 5860129 | 4766 | 0.047 | 0.3794 | No | ||
| 38 | FLNC | 2360048 5050270 | 4791 | 0.046 | 0.3789 | No | ||
| 39 | HOXA3 | 3610632 6020358 | 4940 | 0.042 | 0.3716 | No | ||
| 40 | CAMK2A | 1740333 1940112 4150292 | 5000 | 0.041 | 0.3691 | No | ||
| 41 | HOXB3 | 520292 | 5339 | 0.034 | 0.3513 | No | ||
| 42 | FOXB1 | 4920270 5290463 | 5502 | 0.031 | 0.3431 | No | ||
| 43 | DSG4 | 3990605 6400736 | 5571 | 0.030 | 0.3399 | No | ||
| 44 | RIMS1 | 5670022 | 5901 | 0.025 | 0.3225 | No | ||
| 45 | HOXC4 | 1940193 | 5909 | 0.025 | 0.3225 | No | ||
| 46 | GPR21 | 6520152 | 5952 | 0.024 | 0.3207 | No | ||
| 47 | PROK2 | 540538 | 5965 | 0.024 | 0.3204 | No | ||
| 48 | FZD4 | 1580520 | 6076 | 0.023 | 0.3149 | No | ||
| 49 | TULP1 | 430048 1230156 | 6129 | 0.022 | 0.3124 | No | ||
| 50 | CASK | 2340215 3290576 6770215 | 6245 | 0.021 | 0.3065 | No | ||
| 51 | SLC5A2 | 4480148 6400242 | 6416 | 0.019 | 0.2977 | No | ||
| 52 | CREB5 | 2320368 | 6567 | 0.018 | 0.2898 | No | ||
| 53 | TNFSF4 | 3940594 | 6679 | 0.017 | 0.2841 | No | ||
| 54 | GLRA1 | 610575 | 6722 | 0.017 | 0.2821 | No | ||
| 55 | NEO1 | 1990154 | 6875 | 0.016 | 0.2741 | No | ||
| 56 | VASP | 7050500 | 7056 | 0.014 | 0.2646 | No | ||
| 57 | TYRP1 | 3170450 5690204 6290092 | 7246 | 0.013 | 0.2546 | No | ||
| 58 | HOXC5 | 1580017 | 7434 | 0.012 | 0.2447 | No | ||
| 59 | ERG | 50154 1770739 | 7552 | 0.011 | 0.2385 | No | ||
| 60 | E2F3 | 50162 460180 | 7560 | 0.011 | 0.2383 | No | ||
| 61 | GDF7 | 6370301 | 7619 | 0.011 | 0.2354 | No | ||
| 62 | SLC13A4 | 2940541 | 7664 | 0.011 | 0.2332 | No | ||
| 63 | ESRRG | 4010181 | 7822 | 0.010 | 0.2248 | No | ||
| 64 | EGFL6 | 3940181 | 7891 | 0.010 | 0.2213 | No | ||
| 65 | CTBP2 | 430309 3710079 | 7928 | 0.010 | 0.2195 | No | ||
| 66 | ENTPD1 | 7050139 | 8303 | 0.008 | 0.1994 | No | ||
| 67 | MSX2 | 1570022 | 8343 | 0.008 | 0.1974 | No | ||
| 68 | SLC1A7 | 1990181 | 8344 | 0.008 | 0.1975 | No | ||
| 69 | LIFR | 3360068 3830102 | 8437 | 0.008 | 0.1927 | No | ||
| 70 | OTOP3 | 5360685 | 8537 | 0.007 | 0.1874 | No | ||
| 71 | POU6F2 | 5270451 | 8542 | 0.007 | 0.1873 | No | ||
| 72 | PITX1 | 6550541 | 8655 | 0.007 | 0.1814 | No | ||
| 73 | IRX5 | 2630494 | 8681 | 0.007 | 0.1801 | No | ||
| 74 | BTG4 | 4060020 | 8694 | 0.007 | 0.1796 | No | ||
| 75 | HR | 2690095 6200300 | 8757 | 0.006 | 0.1763 | No | ||
| 76 | PRDM13 | 1430022 | 8963 | 0.006 | 0.1653 | No | ||
| 77 | PCSK1 | 2350148 | 9096 | 0.005 | 0.1582 | No | ||
| 78 | SOX15 | 2470603 | 9140 | 0.005 | 0.1560 | No | ||
| 79 | GRID2 | 2680242 | 9256 | 0.005 | 0.1498 | No | ||
| 80 | LPHN2 | 4480010 | 9376 | 0.004 | 0.1434 | No | ||
| 81 | HOXD10 | 6100039 | 9394 | 0.004 | 0.1426 | No | ||
| 82 | NKX6-1 | 6040731 | 9460 | 0.004 | 0.1391 | No | ||
| 83 | BHLHB5 | 6510520 | 9463 | 0.004 | 0.1391 | No | ||
| 84 | ANGPTL2 | 6980152 6760162 | 9498 | 0.004 | 0.1373 | No | ||
| 85 | SLC12A5 | 1980692 | 9505 | 0.004 | 0.1371 | No | ||
| 86 | GUCY2F | 2450309 | 9605 | 0.004 | 0.1318 | No | ||
| 87 | RTN2 | 4480538 | 9656 | 0.004 | 0.1291 | No | ||
| 88 | DPYD | 6110095 | 9687 | 0.004 | 0.1276 | No | ||
| 89 | NRXN3 | 1400546 3360471 | 9768 | 0.003 | 0.1233 | No | ||
| 90 | SLC6A20 | 5690537 | 9771 | 0.003 | 0.1232 | No | ||
| 91 | PURA | 3870156 | 9895 | 0.003 | 0.1166 | No | ||
| 92 | PAX6 | 1190025 | 9898 | 0.003 | 0.1165 | No | ||
| 93 | AMHR2 | 2810180 3360300 | 9992 | 0.003 | 0.1115 | No | ||
| 94 | DMD | 1740041 3990332 | 10112 | 0.002 | 0.1051 | No | ||
| 95 | MAB21L1 | 1450634 1580092 | 10316 | 0.002 | 0.0941 | No | ||
| 96 | DMRT2 | 2630068 | 10485 | 0.001 | 0.0851 | No | ||
| 97 | PALM2 | 4120068 7100398 | 10531 | 0.001 | 0.0826 | No | ||
| 98 | PITX2 | 870537 2690139 | 10667 | 0.001 | 0.0753 | No | ||
| 99 | BNC2 | 4810603 | 10701 | 0.001 | 0.0735 | No | ||
| 100 | LRRC21 | 990731 | 10822 | 0.000 | 0.0670 | No | ||
| 101 | LIN28 | 6590672 | 10871 | 0.000 | 0.0644 | No | ||
| 102 | SATB2 | 2470181 | 10987 | -0.000 | 0.0582 | No | ||
| 103 | ATRNL1 | 6980368 | 11050 | -0.000 | 0.0548 | No | ||
| 104 | ABCC6 | 6900168 | 11156 | -0.001 | 0.0492 | No | ||
| 105 | MYO3B | 1400092 | 11158 | -0.001 | 0.0491 | No | ||
| 106 | GNB3 | 1230167 | 11170 | -0.001 | 0.0485 | No | ||
| 107 | FOXA1 | 3710609 | 11195 | -0.001 | 0.0472 | No | ||
| 108 | FGF14 | 2630390 3520075 6770048 | 11417 | -0.001 | 0.0353 | No | ||
| 109 | EYA4 | 5360286 | 11418 | -0.001 | 0.0353 | No | ||
| 110 | FGF7 | 5390484 | 11438 | -0.002 | 0.0343 | No | ||
| 111 | SST | 6590142 | 11525 | -0.002 | 0.0297 | No | ||
| 112 | EFS | 110288 | 11788 | -0.003 | 0.0155 | No | ||
| 113 | FLRT1 | 4540348 | 11835 | -0.003 | 0.0131 | No | ||
| 114 | GRIN2B | 3800333 | 11884 | -0.003 | 0.0105 | No | ||
| 115 | CRYGS | 2680398 | 11955 | -0.003 | 0.0068 | No | ||
| 116 | RAX | 4590600 | 11965 | -0.003 | 0.0063 | No | ||
| 117 | ATP6V0A4 | 360100 | 12015 | -0.004 | 0.0037 | No | ||
| 118 | SIX3 | 3830402 | 12017 | -0.004 | 0.0038 | No | ||
| 119 | NLGN1 | 5670278 | 12288 | -0.005 | -0.0108 | No | ||
| 120 | NR1H4 | 2450176 | 12390 | -0.005 | -0.0162 | No | ||
| 121 | GAP43 | 3130504 | 12528 | -0.006 | -0.0235 | No | ||
| 122 | ERBB4 | 4610400 4810017 | 12582 | -0.006 | -0.0263 | No | ||
| 123 | TCF1 | 5390022 | 12587 | -0.006 | -0.0264 | No | ||
| 124 | AGA | 6350292 | 12632 | -0.006 | -0.0287 | No | ||
| 125 | PAX7 | 6860593 | 12693 | -0.006 | -0.0319 | No | ||
| 126 | TLX3 | 510619 | 13035 | -0.008 | -0.0502 | No | ||
| 127 | KCNIP2 | 60088 1780324 | 13110 | -0.009 | -0.0541 | No | ||
| 128 | SLC6A5 | 870121 | 13124 | -0.009 | -0.0546 | No | ||
| 129 | JUB | 5860711 5900500 | 13180 | -0.009 | -0.0574 | No | ||
| 130 | ADAMTSL1 | 2970092 | 13249 | -0.010 | -0.0610 | No | ||
| 131 | WNT3A | 6350348 | 13276 | -0.010 | -0.0622 | No | ||
| 132 | MEOX2 | 6400288 | 13392 | -0.011 | -0.0682 | No | ||
| 133 | JARID2 | 6290538 | 13395 | -0.011 | -0.0682 | No | ||
| 134 | MEIS1 | 1400575 | 13430 | -0.012 | -0.0698 | No | ||
| 135 | MDGA1 | 1170139 | 13439 | -0.012 | -0.0700 | No | ||
| 136 | CLDN2 | 580053 | 13507 | -0.012 | -0.0735 | No | ||
| 137 | PRRX1 | 4120193 4480390 | 13546 | -0.013 | -0.0753 | No | ||
| 138 | LEMD1 | 3990132 5720187 | 13759 | -0.015 | -0.0865 | No | ||
| 139 | PDE4D | 2470528 6660014 | 13911 | -0.017 | -0.0944 | No | ||
| 140 | ROBO3 | 6550408 | 13920 | -0.017 | -0.0946 | No | ||
| 141 | STAT5B | 6200026 | 13947 | -0.018 | -0.0957 | No | ||
| 142 | MOAP1 | 580537 | 14173 | -0.022 | -0.1075 | No | ||
| 143 | GPX1 | 4150093 | 14266 | -0.024 | -0.1121 | No | ||
| 144 | NOTCH2 | 2570397 | 14297 | -0.025 | -0.1133 | No | ||
| 145 | SOAT2 | 1230324 3610603 | 14386 | -0.028 | -0.1176 | No | ||
| 146 | TNFSF11 | 6350204 | 14396 | -0.028 | -0.1176 | No | ||
| 147 | JMJD1C | 940575 2120025 | 14430 | -0.029 | -0.1189 | No | ||
| 148 | PSCD2 | 3870113 | 14501 | -0.032 | -0.1221 | No | ||
| 149 | AKAP2 | 5550347 | 14757 | -0.044 | -0.1352 | No | ||
| 150 | ZNF385 | 450176 4810358 | 14910 | -0.053 | -0.1425 | No | ||
| 151 | THRAP1 | 2470121 | 15024 | -0.059 | -0.1476 | No | ||
| 152 | ABLIM1 | 580170 3710338 6520504 | 15171 | -0.069 | -0.1544 | No | ||
| 153 | RORA | 380164 5220215 | 15187 | -0.071 | -0.1540 | No | ||
| 154 | LTBP1 | 780035 2100746 2760402 3780551 6550184 6860193 | 15247 | -0.075 | -0.1559 | No | ||
| 155 | CLASP1 | 6860279 | 15375 | -0.088 | -0.1612 | No | ||
| 156 | NCOA2 | 940347 2850725 | 15436 | -0.094 | -0.1629 | No | ||
| 157 | IGSF3 | 6450524 | 15555 | -0.107 | -0.1674 | No | ||
| 158 | LDB2 | 5670441 6110170 | 15830 | -0.138 | -0.1799 | No | ||
| 159 | TCF7L2 | 2190348 5220113 | 15855 | -0.142 | -0.1788 | No | ||
| 160 | CNOT4 | 2320242 6760706 | 15894 | -0.148 | -0.1783 | No | ||
| 161 | BTRC | 3170131 | 16035 | -0.169 | -0.1830 | No | ||
| 162 | NR6A1 | 4010347 | 16300 | -0.216 | -0.1936 | No | ||
| 163 | RG9MTD2 | 6520100 | 16414 | -0.239 | -0.1956 | No | ||
| 164 | AMMECR1 | 3800435 | 16463 | -0.254 | -0.1939 | No | ||
| 165 | CHORDC1 | 6650577 | 16499 | -0.264 | -0.1912 | No | ||
| 166 | BCL11A | 6860369 | 16672 | -0.312 | -0.1952 | No | ||
| 167 | TIGD2 | 7100129 | 16682 | -0.315 | -0.1903 | No | ||
| 168 | CHD2 | 4070039 | 17007 | -0.417 | -0.2007 | No | ||
| 169 | DCTN1 | 1450056 6590022 | 17070 | -0.437 | -0.1965 | No | ||
| 170 | NFIX | 2450152 | 17139 | -0.468 | -0.1921 | No | ||
| 171 | HIST1H1C | 3870603 | 17181 | -0.484 | -0.1860 | No | ||
| 172 | ATP1B3 | 6040128 | 17549 | -0.657 | -0.1946 | No | ||
| 173 | SRPK2 | 6380341 | 17799 | -0.805 | -0.1943 | No | ||
| 174 | RPA3 | 5700136 | 17876 | -0.871 | -0.1834 | No | ||
| 175 | DEF6 | 840593 | 18019 | -1.006 | -0.1738 | No | ||
| 176 | SSBP2 | 2260725 2900400 4730711 | 18425 | -1.846 | -0.1640 | No | ||
| 177 | KCNH2 | 1170451 | 18487 | -2.154 | -0.1302 | No | ||
| 178 | SATB1 | 5670154 | 18523 | -2.380 | -0.0912 | No | ||
| 179 | TLE4 | 1450064 1500519 | 18569 | -2.751 | -0.0463 | No | ||
| 180 | ETS2 | 360451 | 18573 | -2.835 | 0.0023 | No |

