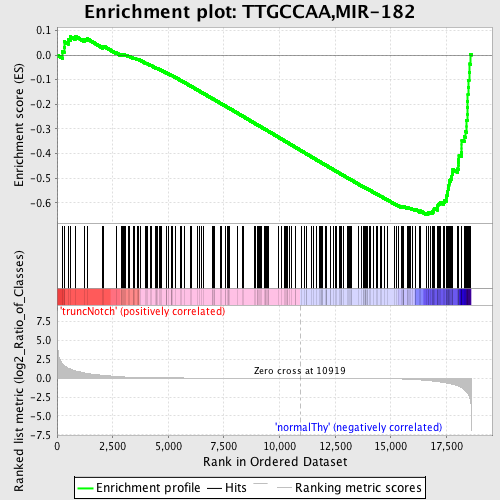
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | TTGCCAA,MIR-182 |
| Enrichment Score (ES) | -0.6469418 |
| Normalized Enrichment Score (NES) | -1.7727169 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.010694041 |
| FWER p-Value | 0.01 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RTN4 | 50725 630647 870097 870184 | 236 | 1.892 | 0.0135 | No | ||
| 2 | NCALD | 730176 | 316 | 1.667 | 0.0325 | No | ||
| 3 | AUP1 | 4150075 6980551 | 338 | 1.608 | 0.0537 | No | ||
| 4 | CFL1 | 2340735 | 510 | 1.318 | 0.0628 | No | ||
| 5 | PPIL1 | 6350435 | 604 | 1.187 | 0.0743 | No | ||
| 6 | PPP3R1 | 1190041 1570487 | 810 | 0.980 | 0.0768 | No | ||
| 7 | LIMK1 | 5080064 | 1220 | 0.704 | 0.0644 | No | ||
| 8 | ARHGEF3 | 3440156 | 1347 | 0.630 | 0.0663 | No | ||
| 9 | CORO1C | 4570397 | 2058 | 0.379 | 0.0330 | No | ||
| 10 | DENR | 4120176 | 2087 | 0.372 | 0.0366 | No | ||
| 11 | P2RX4 | 2060497 6650162 | 2656 | 0.227 | 0.0089 | No | ||
| 12 | TEX2 | 1740504 2480102 3060494 | 2876 | 0.184 | -0.0004 | No | ||
| 13 | SNAP23 | 2480722 5390315 | 2887 | 0.183 | 0.0016 | No | ||
| 14 | ACACA | 2490612 2680369 | 2921 | 0.176 | 0.0022 | No | ||
| 15 | SS18L1 | 4610180 6550068 | 2988 | 0.167 | 0.0010 | No | ||
| 16 | EVI5 | 6380040 | 3021 | 0.163 | 0.0015 | No | ||
| 17 | EGR3 | 6940128 | 3087 | 0.153 | 0.0001 | No | ||
| 18 | SUHW2 | 5290192 | 3221 | 0.137 | -0.0052 | No | ||
| 19 | PYGO2 | 360494 | 3247 | 0.134 | -0.0047 | No | ||
| 20 | SEPT9 | 1940040 | 3441 | 0.114 | -0.0136 | No | ||
| 21 | BCL2L12 | 5360176 | 3448 | 0.114 | -0.0124 | No | ||
| 22 | KDELR1 | 6760504 | 3476 | 0.110 | -0.0123 | No | ||
| 23 | SMAD1 | 630537 1850333 | 3620 | 0.099 | -0.0187 | No | ||
| 24 | MYOHD1 | 1410520 | 3640 | 0.098 | -0.0184 | No | ||
| 25 | EPAS1 | 5290156 | 3671 | 0.095 | -0.0187 | No | ||
| 26 | INSIG1 | 1500332 3450458 4540301 | 3752 | 0.090 | -0.0218 | No | ||
| 27 | EIF3S10 | 1190079 3840176 | 3966 | 0.078 | -0.0323 | No | ||
| 28 | FBXW7 | 4210338 7050280 | 4001 | 0.076 | -0.0331 | No | ||
| 29 | BCL2L13 | 3170593 | 4041 | 0.074 | -0.0341 | No | ||
| 30 | YAF2 | 940053 | 4218 | 0.066 | -0.0428 | No | ||
| 31 | UBXD3 | 5050193 | 4220 | 0.066 | -0.0419 | No | ||
| 32 | CTTN | 780154 1690538 3310017 4060717 | 4411 | 0.058 | -0.0515 | No | ||
| 33 | NTNG1 | 50270 1410427 4810333 | 4464 | 0.056 | -0.0535 | No | ||
| 34 | HOXA10 | 6110397 7100458 | 4487 | 0.056 | -0.0539 | No | ||
| 35 | PDZD4 | 840528 6290438 6350044 | 4508 | 0.055 | -0.0542 | No | ||
| 36 | KPNA3 | 7040088 | 4609 | 0.052 | -0.0589 | No | ||
| 37 | THBS2 | 2850136 | 4648 | 0.051 | -0.0603 | No | ||
| 38 | MYADM | 4230132 | 4711 | 0.049 | -0.0630 | No | ||
| 39 | PTHLH | 5290739 | 4894 | 0.044 | -0.0723 | No | ||
| 40 | ABCC3 | 4050242 | 4896 | 0.044 | -0.0717 | No | ||
| 41 | ADCY6 | 450364 6290670 6940286 | 4929 | 0.043 | -0.0729 | No | ||
| 42 | EDNRB | 4280717 6400435 | 5001 | 0.041 | -0.0762 | No | ||
| 43 | ACVR1 | 6840671 | 5119 | 0.039 | -0.0820 | No | ||
| 44 | RDX | 4200039 6550465 | 5143 | 0.038 | -0.0827 | No | ||
| 45 | CITED2 | 5670114 5130088 | 5161 | 0.038 | -0.0831 | No | ||
| 46 | GABBR1 | 2850519 2850735 | 5169 | 0.038 | -0.0829 | No | ||
| 47 | CEBPA | 5690519 | 5330 | 0.035 | -0.0912 | No | ||
| 48 | FGF9 | 1050195 | 5564 | 0.030 | -0.1034 | No | ||
| 49 | RHOBTB1 | 1410010 | 5568 | 0.030 | -0.1032 | No | ||
| 50 | VLDLR | 870722 3060047 5340452 6550131 | 5740 | 0.027 | -0.1121 | No | ||
| 51 | ZMPSTE24 | 1090112 2030593 3780129 4590044 | 5991 | 0.024 | -0.1253 | No | ||
| 52 | RALGDS | 430463 | 6045 | 0.023 | -0.1279 | No | ||
| 53 | HMGCLL1 | 2570091 | 6328 | 0.020 | -0.1429 | No | ||
| 54 | NFASC | 1940619 2470156 4590168 6840524 | 6408 | 0.019 | -0.1470 | No | ||
| 55 | CD2AP | 1940369 | 6506 | 0.019 | -0.1520 | No | ||
| 56 | VAMP3 | 7100050 | 6559 | 0.018 | -0.1546 | No | ||
| 57 | APLN | 6620632 | 6982 | 0.015 | -0.1773 | No | ||
| 58 | BRUNOL6 | 6840373 | 7004 | 0.015 | -0.1782 | No | ||
| 59 | ADCY2 | 4610100 | 7007 | 0.015 | -0.1781 | No | ||
| 60 | VASP | 7050500 | 7056 | 0.014 | -0.1806 | No | ||
| 61 | TXNL1 | 2850148 | 7061 | 0.014 | -0.1806 | No | ||
| 62 | DAZAP2 | 6200102 | 7352 | 0.012 | -0.1962 | No | ||
| 63 | DCUN1D1 | 2360332 | 7372 | 0.012 | -0.1970 | No | ||
| 64 | ARRDC3 | 4610390 | 7551 | 0.011 | -0.2065 | No | ||
| 65 | NKX2-2 | 4150731 | 7557 | 0.011 | -0.2067 | No | ||
| 66 | HOXA9 | 610494 4730040 5130601 | 7639 | 0.011 | -0.2109 | No | ||
| 67 | EPHB1 | 4610056 | 7721 | 0.011 | -0.2152 | No | ||
| 68 | DSCAM | 1780050 2450731 2810438 | 7722 | 0.010 | -0.2150 | No | ||
| 69 | SLC1A2 | 2230520 | 7755 | 0.010 | -0.2166 | No | ||
| 70 | PAFAH1B1 | 4230333 6420121 6450066 | 8128 | 0.009 | -0.2367 | No | ||
| 71 | ARMC1 | 5220458 | 8320 | 0.008 | -0.2470 | No | ||
| 72 | SOX2 | 630487 2230491 | 8394 | 0.008 | -0.2509 | No | ||
| 73 | ADRA2C | 5130154 | 8884 | 0.006 | -0.2774 | No | ||
| 74 | GMFB | 6590035 | 8911 | 0.006 | -0.2787 | No | ||
| 75 | EBF3 | 4200129 | 9008 | 0.005 | -0.2838 | No | ||
| 76 | PRKD1 | 1410706 5090438 | 9023 | 0.005 | -0.2845 | No | ||
| 77 | IBRDC2 | 5130041 | 9073 | 0.005 | -0.2871 | No | ||
| 78 | BHLHB3 | 3450438 | 9088 | 0.005 | -0.2878 | No | ||
| 79 | ANK2 | 6510546 | 9151 | 0.005 | -0.2911 | No | ||
| 80 | FMR1 | 5050075 | 9172 | 0.005 | -0.2921 | No | ||
| 81 | MAK | 2940706 | 9330 | 0.005 | -0.3006 | No | ||
| 82 | EIF2C1 | 6900551 | 9369 | 0.004 | -0.3026 | No | ||
| 83 | LPHN2 | 4480010 | 9376 | 0.004 | -0.3029 | No | ||
| 84 | PCDH8 | 2360047 4200452 | 9421 | 0.004 | -0.3052 | No | ||
| 85 | BHLHB5 | 6510520 | 9463 | 0.004 | -0.3074 | No | ||
| 86 | CHES1 | 4570121 5700162 | 9508 | 0.004 | -0.3097 | No | ||
| 87 | ADAM10 | 3780156 | 9517 | 0.004 | -0.3101 | No | ||
| 88 | QKI | 5220093 6130044 | 9948 | 0.003 | -0.3334 | No | ||
| 89 | PLD1 | 2450537 | 10080 | 0.002 | -0.3405 | No | ||
| 90 | GRINL1A | 6350128 | 10216 | 0.002 | -0.3478 | No | ||
| 91 | PCDH18 | 430010 | 10284 | 0.002 | -0.3514 | No | ||
| 92 | SCRT1 | 6760736 | 10305 | 0.002 | -0.3525 | No | ||
| 93 | MMP16 | 2680139 | 10370 | 0.002 | -0.3560 | No | ||
| 94 | MEF2C | 670025 780338 | 10376 | 0.002 | -0.3562 | No | ||
| 95 | SEPT7 | 2760685 | 10426 | 0.001 | -0.3589 | No | ||
| 96 | EPHA7 | 1190288 4010278 | 10549 | 0.001 | -0.3655 | No | ||
| 97 | ANUBL1 | 1580113 | 10696 | 0.001 | -0.3734 | No | ||
| 98 | BNC2 | 4810603 | 10701 | 0.001 | -0.3736 | No | ||
| 99 | SATB2 | 2470181 | 10987 | -0.000 | -0.3891 | No | ||
| 100 | SH3BP4 | 3140242 | 11131 | -0.001 | -0.3969 | No | ||
| 101 | KLF15 | 4230164 | 11199 | -0.001 | -0.4005 | No | ||
| 102 | BDNF | 2940128 3520368 | 11224 | -0.001 | -0.4018 | No | ||
| 103 | RIMS3 | 3890563 | 11414 | -0.001 | -0.4121 | No | ||
| 104 | ADAMTS18 | 5690008 | 11507 | -0.002 | -0.4170 | No | ||
| 105 | RGS17 | 1400603 | 11671 | -0.002 | -0.4259 | No | ||
| 106 | PRDM1 | 3170347 3520301 | 11775 | -0.003 | -0.4314 | No | ||
| 107 | NPTX1 | 3710736 | 11851 | -0.003 | -0.4355 | No | ||
| 108 | ELL2 | 6420091 | 11886 | -0.003 | -0.4373 | No | ||
| 109 | FOXF2 | 1240091 | 11898 | -0.003 | -0.4378 | No | ||
| 110 | STARD13 | 540722 | 11912 | -0.003 | -0.4385 | No | ||
| 111 | ZCCHC14 | 5900288 | 11936 | -0.003 | -0.4397 | No | ||
| 112 | CUL5 | 450142 | 12053 | -0.004 | -0.4459 | No | ||
| 113 | NPM1 | 4730427 | 12090 | -0.004 | -0.4478 | No | ||
| 114 | ATOH8 | 4540204 | 12115 | -0.004 | -0.4491 | No | ||
| 115 | CLOCK | 1410070 2940292 3390075 | 12130 | -0.004 | -0.4498 | No | ||
| 116 | SP3 | 3840338 | 12266 | -0.004 | -0.4571 | No | ||
| 117 | RET | 3850746 | 12283 | -0.005 | -0.4579 | No | ||
| 118 | GRM5 | 60528 | 12421 | -0.005 | -0.4653 | No | ||
| 119 | CCNJ | 870047 | 12505 | -0.006 | -0.4697 | No | ||
| 120 | PRPF4B | 940181 2570300 | 12536 | -0.006 | -0.4713 | No | ||
| 121 | MTSS1 | 780435 2370114 | 12710 | -0.006 | -0.4806 | No | ||
| 122 | FZD3 | 1500397 | 12737 | -0.007 | -0.4819 | No | ||
| 123 | UBE2R2 | 1230504 | 12798 | -0.007 | -0.4851 | No | ||
| 124 | EFNB2 | 5340136 | 12804 | -0.007 | -0.4852 | No | ||
| 125 | TMEM47 | 1660086 | 12866 | -0.007 | -0.4884 | No | ||
| 126 | RARG | 6760136 | 13031 | -0.008 | -0.4972 | No | ||
| 127 | FBXW11 | 6450632 | 13100 | -0.009 | -0.5008 | No | ||
| 128 | ARHGEF7 | 1690441 | 13144 | -0.009 | -0.5030 | No | ||
| 129 | JUB | 5860711 5900500 | 13180 | -0.009 | -0.5048 | No | ||
| 130 | MITF | 380056 | 13233 | -0.010 | -0.5075 | No | ||
| 131 | AKAP7 | 4610044 | 13251 | -0.010 | -0.5083 | No | ||
| 132 | PRRX1 | 4120193 4480390 | 13546 | -0.013 | -0.5241 | No | ||
| 133 | PHF21A | 610014 | 13678 | -0.014 | -0.5310 | No | ||
| 134 | ISL1 | 4810239 | 13779 | -0.016 | -0.5362 | No | ||
| 135 | EPM2A | 2640079 | 13782 | -0.016 | -0.5361 | No | ||
| 136 | L1CAM | 4850021 | 13818 | -0.016 | -0.5378 | No | ||
| 137 | TOPBP1 | 6020333 | 13849 | -0.016 | -0.5392 | No | ||
| 138 | BRE | 2340273 3190731 4050500 | 13855 | -0.016 | -0.5393 | No | ||
| 139 | DR1 | 5860750 | 13864 | -0.016 | -0.5395 | No | ||
| 140 | ELMO1 | 110338 1190451 1580148 1850609 2350706 2370369 2450129 3840113 4150184 4280079 6290066 6380435 6770494 7100180 | 13922 | -0.018 | -0.5423 | No | ||
| 141 | RAB6B | 5860121 | 13950 | -0.018 | -0.5435 | No | ||
| 142 | BTBD11 | 6650154 | 13971 | -0.018 | -0.5444 | No | ||
| 143 | LHX3 | 670164 1770280 | 14032 | -0.019 | -0.5474 | No | ||
| 144 | SLITRK4 | 5910022 | 14088 | -0.020 | -0.5501 | No | ||
| 145 | ARF4 | 3190661 | 14224 | -0.023 | -0.5571 | No | ||
| 146 | KHDRBS3 | 2120154 | 14377 | -0.027 | -0.5650 | No | ||
| 147 | ANK3 | 1500161 2900435 4210154 6400100 | 14419 | -0.029 | -0.5668 | No | ||
| 148 | MEF2D | 5690576 | 14536 | -0.033 | -0.5726 | No | ||
| 149 | PIGA | 1940435 5290692 | 14563 | -0.034 | -0.5736 | No | ||
| 150 | XPR1 | 1500093 | 14736 | -0.043 | -0.5823 | No | ||
| 151 | NUMB | 2450735 3800253 6350040 | 14867 | -0.050 | -0.5887 | No | ||
| 152 | ABLIM1 | 580170 3710338 6520504 | 15171 | -0.069 | -0.6042 | No | ||
| 153 | KTN1 | 70446 3450609 4560048 | 15261 | -0.076 | -0.6080 | No | ||
| 154 | DOK4 | 4060452 | 15352 | -0.085 | -0.6117 | No | ||
| 155 | SPRY4 | 1570594 | 15480 | -0.099 | -0.6172 | No | ||
| 156 | GPR85 | 5130750 5910020 | 15532 | -0.103 | -0.6186 | No | ||
| 157 | ARHGDIA | 5900239 | 15554 | -0.107 | -0.6182 | No | ||
| 158 | IGSF3 | 6450524 | 15555 | -0.107 | -0.6167 | No | ||
| 159 | RAB10 | 2360008 5890020 | 15563 | -0.108 | -0.6156 | No | ||
| 160 | MBNL2 | 3450707 | 15577 | -0.109 | -0.6148 | No | ||
| 161 | ADD3 | 4150739 5390600 | 15732 | -0.127 | -0.6214 | No | ||
| 162 | SLC39A9 | 3940348 | 15737 | -0.128 | -0.6198 | No | ||
| 163 | MAGI1 | 6510500 1990239 4670471 | 15774 | -0.131 | -0.6200 | No | ||
| 164 | VAV2 | 3610725 5890717 | 15847 | -0.140 | -0.6219 | No | ||
| 165 | ELL | 3520731 | 15885 | -0.146 | -0.6219 | No | ||
| 166 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 15995 | -0.163 | -0.6256 | No | ||
| 167 | ATXN1 | 5550156 | 16088 | -0.176 | -0.6281 | No | ||
| 168 | RAC1 | 4810687 | 16093 | -0.176 | -0.6259 | No | ||
| 169 | NAP1L1 | 2510593 5900215 5090470 450079 4920280 | 16308 | -0.218 | -0.6345 | No | ||
| 170 | SNX4 | 1980358 | 16330 | -0.222 | -0.6325 | No | ||
| 171 | AEBP2 | 2970603 | 16594 | -0.292 | -0.6428 | Yes | ||
| 172 | BCL11A | 6860369 | 16672 | -0.312 | -0.6426 | Yes | ||
| 173 | EXOC4 | 2470253 | 16689 | -0.317 | -0.6390 | Yes | ||
| 174 | WDR40A | 2190167 | 16771 | -0.341 | -0.6387 | Yes | ||
| 175 | SLC4A7 | 2370056 | 16858 | -0.368 | -0.6382 | Yes | ||
| 176 | MTCH2 | 3780603 6620215 | 16879 | -0.372 | -0.6342 | Yes | ||
| 177 | MMD | 3850239 | 16934 | -0.389 | -0.6317 | Yes | ||
| 178 | YWHAG | 3780341 | 16937 | -0.391 | -0.6263 | Yes | ||
| 179 | EIF5 | 6380750 | 16967 | -0.403 | -0.6223 | Yes | ||
| 180 | PPP1R2 | 3710278 1400725 | 17093 | -0.444 | -0.6229 | Yes | ||
| 181 | PAIP2 | 4210441 | 17095 | -0.444 | -0.6168 | Yes | ||
| 182 | CDV3 | 1940577 | 17114 | -0.457 | -0.6114 | Yes | ||
| 183 | SYPL1 | 2680706 5900403 | 17124 | -0.463 | -0.6054 | Yes | ||
| 184 | KCMF1 | 2970577 | 17179 | -0.484 | -0.6016 | Yes | ||
| 185 | MFAP3 | 6380066 6520487 | 17235 | -0.506 | -0.5976 | Yes | ||
| 186 | PHF13 | 450435 | 17378 | -0.569 | -0.5973 | Yes | ||
| 187 | DOCK9 | 5080037 | 17413 | -0.584 | -0.5911 | Yes | ||
| 188 | DYNC1LI2 | 50338 | 17489 | -0.626 | -0.5864 | Yes | ||
| 189 | ATP2A2 | 1090075 3990279 | 17491 | -0.626 | -0.5777 | Yes | ||
| 190 | FEM1C | 60300 6130717 6980164 | 17520 | -0.641 | -0.5703 | Yes | ||
| 191 | GIT2 | 6040039 6770181 | 17540 | -0.652 | -0.5623 | Yes | ||
| 192 | ATP1B3 | 6040128 | 17549 | -0.657 | -0.5536 | Yes | ||
| 193 | BAG4 | 2260070 | 17570 | -0.668 | -0.5453 | Yes | ||
| 194 | PCMT1 | 4590338 4590541 | 17594 | -0.680 | -0.5371 | Yes | ||
| 195 | TSNAX | 3450279 | 17608 | -0.689 | -0.5282 | Yes | ||
| 196 | MAP1LC3B | 730142 | 17615 | -0.691 | -0.5189 | Yes | ||
| 197 | WASF2 | 4850592 | 17631 | -0.698 | -0.5100 | Yes | ||
| 198 | RASA1 | 1240315 | 17696 | -0.741 | -0.5032 | Yes | ||
| 199 | OGT | 2360131 4610333 | 17729 | -0.756 | -0.4944 | Yes | ||
| 200 | BACH2 | 6760131 | 17749 | -0.775 | -0.4846 | Yes | ||
| 201 | BRMS1L | 6110609 | 17778 | -0.791 | -0.4751 | Yes | ||
| 202 | IHPK1 | 1340053 | 17788 | -0.797 | -0.4645 | Yes | ||
| 203 | TAF15 | 4120736 | 17999 | -0.981 | -0.4622 | Yes | ||
| 204 | ZNRF1 | 70592 | 18032 | -1.019 | -0.4498 | Yes | ||
| 205 | TRIM8 | 6130441 | 18037 | -1.028 | -0.4357 | Yes | ||
| 206 | DYRK1A | 3190181 | 18053 | -1.036 | -0.4221 | Yes | ||
| 207 | TOB1 | 4150138 | 18063 | -1.050 | -0.4079 | Yes | ||
| 208 | FLOT1 | 1660575 1690040 | 18166 | -1.206 | -0.3967 | Yes | ||
| 209 | MARCKS | 7040450 | 18194 | -1.252 | -0.3807 | Yes | ||
| 210 | WSB1 | 870563 | 18195 | -1.252 | -0.3632 | Yes | ||
| 211 | MARK3 | 2370450 | 18196 | -1.253 | -0.3458 | Yes | ||
| 212 | KLF7 | 50390 | 18299 | -1.477 | -0.3308 | Yes | ||
| 213 | PRKACB | 4210170 | 18351 | -1.640 | -0.3107 | Yes | ||
| 214 | PBX2 | 3130433 6510292 | 18400 | -1.770 | -0.2886 | Yes | ||
| 215 | KLF13 | 4670563 | 18402 | -1.779 | -0.2639 | Yes | ||
| 216 | RNF44 | 5910692 | 18432 | -1.871 | -0.2394 | Yes | ||
| 217 | CD47 | 1850603 | 18439 | -1.885 | -0.2135 | Yes | ||
| 218 | XBP1 | 3840594 | 18452 | -1.925 | -0.1873 | Yes | ||
| 219 | UBE3C | 3140673 | 18466 | -2.024 | -0.1598 | Yes | ||
| 220 | ZFP36L1 | 2510138 4120048 | 18473 | -2.052 | -0.1315 | Yes | ||
| 221 | CSNK1E | 2850347 5050093 6110301 | 18491 | -2.187 | -0.1020 | Yes | ||
| 222 | SH2D1A | 4760025 | 18516 | -2.349 | -0.0706 | Yes | ||
| 223 | ZFP36 | 2030605 | 18555 | -2.611 | -0.0362 | Yes | ||
| 224 | ETS2 | 360451 | 18573 | -2.835 | 0.0023 | Yes |

