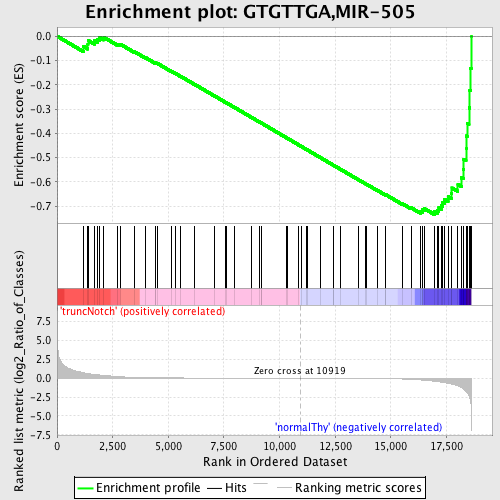
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | GTGTTGA,MIR-505 |
| Enrichment Score (ES) | -0.7343938 |
| Normalized Enrichment Score (NES) | -1.7217923 |
| Nominal p-value | 0.002631579 |
| FDR q-value | 0.007173597 |
| FWER p-Value | 0.027 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC25A3 | 670112 | 1180 | 0.735 | -0.0421 | No | ||
| 2 | PMP22 | 4010239 6550072 | 1383 | 0.615 | -0.0350 | No | ||
| 3 | ALS2 | 3130546 6400070 | 1389 | 0.612 | -0.0174 | No | ||
| 4 | B4GALT1 | 6980167 | 1674 | 0.497 | -0.0181 | No | ||
| 5 | CAMK2D | 2370685 | 1824 | 0.447 | -0.0131 | No | ||
| 6 | SIRT3 | 4010440 6040431 | 1919 | 0.423 | -0.0058 | No | ||
| 7 | DDX3X | 2190020 | 2105 | 0.368 | -0.0050 | No | ||
| 8 | ZMYND11 | 630181 2570019 4850138 5360040 | 2730 | 0.209 | -0.0325 | No | ||
| 9 | SON | 3120148 5860239 7040403 | 2868 | 0.186 | -0.0344 | No | ||
| 10 | MXI1 | 5050064 5130484 | 3466 | 0.111 | -0.0634 | No | ||
| 11 | EIF3S10 | 1190079 3840176 | 3966 | 0.078 | -0.0880 | No | ||
| 12 | EPN2 | 1340451 6550239 6860020 6860403 | 4414 | 0.058 | -0.1104 | No | ||
| 13 | YTHDF3 | 4540632 4670717 7040609 | 4429 | 0.057 | -0.1095 | No | ||
| 14 | GCC1 | 2510132 | 4490 | 0.056 | -0.1111 | No | ||
| 15 | CITED2 | 5670114 5130088 | 5161 | 0.038 | -0.1461 | No | ||
| 16 | GLP1R | 6420528 | 5308 | 0.035 | -0.1530 | No | ||
| 17 | CANX | 4760402 | 5561 | 0.030 | -0.1657 | No | ||
| 18 | SLC18A2 | 6420541 | 6159 | 0.022 | -0.1972 | No | ||
| 19 | ELAVL2 | 360181 | 7089 | 0.014 | -0.2469 | No | ||
| 20 | FZD8 | 1990053 | 7567 | 0.011 | -0.2723 | No | ||
| 21 | SRGAP2 | 1050092 1230403 3830066 6130215 | 7597 | 0.011 | -0.2735 | No | ||
| 22 | HNT | 6450731 | 7985 | 0.009 | -0.2941 | No | ||
| 23 | DPYSL3 | 1580594 | 8752 | 0.006 | -0.3352 | No | ||
| 24 | CUGBP1 | 450292 510022 7050176 7050215 | 9112 | 0.005 | -0.3544 | No | ||
| 25 | CYP46A1 | 2470332 | 9179 | 0.005 | -0.3578 | No | ||
| 26 | STRN3 | 1450093 6200685 | 9199 | 0.005 | -0.3587 | No | ||
| 27 | ZIC2 | 1410408 | 10312 | 0.002 | -0.4186 | No | ||
| 28 | FIGN | 7000601 | 10364 | 0.002 | -0.4213 | No | ||
| 29 | LIN28 | 6590672 | 10871 | 0.000 | -0.4486 | No | ||
| 30 | KCNJ2 | 630019 | 11004 | -0.000 | -0.4557 | No | ||
| 31 | MLLT7 | 4480707 | 11196 | -0.001 | -0.4660 | No | ||
| 32 | AKAP1 | 110148 1740735 2260019 7000563 | 11256 | -0.001 | -0.4691 | No | ||
| 33 | GDA | 3290435 | 11839 | -0.003 | -0.5004 | No | ||
| 34 | MAFB | 1230471 | 12410 | -0.005 | -0.5310 | No | ||
| 35 | ATP2B2 | 3780397 | 12439 | -0.005 | -0.5323 | No | ||
| 36 | USP47 | 2640497 | 12755 | -0.007 | -0.5491 | No | ||
| 37 | PCDH9 | 2100136 | 13555 | -0.013 | -0.5918 | No | ||
| 38 | DDX3Y | 1580278 4200519 | 13858 | -0.016 | -0.6076 | No | ||
| 39 | PDE4D | 2470528 6660014 | 13911 | -0.017 | -0.6099 | No | ||
| 40 | MBNL1 | 2640762 7100048 | 14387 | -0.028 | -0.6347 | No | ||
| 41 | AKAP2 | 5550347 | 14757 | -0.044 | -0.6533 | No | ||
| 42 | CREB1 | 1500717 2230358 3610600 6550601 | 14774 | -0.045 | -0.6529 | No | ||
| 43 | GPR85 | 5130750 5910020 | 15532 | -0.103 | -0.6907 | No | ||
| 44 | UBE2D3 | 3190452 | 15912 | -0.149 | -0.7067 | No | ||
| 45 | RTF1 | 2100735 | 16338 | -0.223 | -0.7231 | Yes | ||
| 46 | BICD2 | 1090411 | 16418 | -0.240 | -0.7204 | Yes | ||
| 47 | HIPK1 | 110193 | 16427 | -0.243 | -0.7137 | Yes | ||
| 48 | SLC12A2 | 4610019 6040672 | 16504 | -0.265 | -0.7100 | Yes | ||
| 49 | MKRN1 | 1410097 | 16957 | -0.400 | -0.7227 | Yes | ||
| 50 | KHDRBS1 | 1240403 6040040 | 17088 | -0.442 | -0.7168 | Yes | ||
| 51 | KRAS | 2060170 | 17160 | -0.476 | -0.7067 | Yes | ||
| 52 | MECP2 | 1940450 | 17281 | -0.523 | -0.6978 | Yes | ||
| 53 | CTDSPL | 4670546 | 17333 | -0.549 | -0.6845 | Yes | ||
| 54 | MAPK14 | 5290731 | 17410 | -0.583 | -0.6716 | Yes | ||
| 55 | H2AFV | 2030577 5220129 | 17573 | -0.670 | -0.6607 | Yes | ||
| 56 | RANBP9 | 4670685 | 17730 | -0.757 | -0.6470 | Yes | ||
| 57 | DHX15 | 870632 | 17748 | -0.775 | -0.6252 | Yes | ||
| 58 | CGGBP1 | 4810053 | 18017 | -1.005 | -0.6103 | Yes | ||
| 59 | NFE2L1 | 6130053 | 18175 | -1.214 | -0.5832 | Yes | ||
| 60 | RUNX1 | 3840711 | 18253 | -1.370 | -0.5473 | Yes | ||
| 61 | SFRS1 | 2360440 | 18281 | -1.429 | -0.5069 | Yes | ||
| 62 | ARNTL | 3170463 | 18381 | -1.727 | -0.4617 | Yes | ||
| 63 | PDPK1 | 6650168 | 18388 | -1.747 | -0.4109 | Yes | ||
| 64 | RNF44 | 5910692 | 18432 | -1.871 | -0.3585 | Yes | ||
| 65 | BCL11B | 2680673 | 18514 | -2.342 | -0.2943 | Yes | ||
| 66 | CNIH | 2190484 5080609 | 18550 | -2.537 | -0.2220 | Yes | ||
| 67 | CYFIP2 | 520577 6040300 | 18590 | -3.102 | -0.1333 | Yes | ||
| 68 | HRBL | 1850102 3830253 4780176 | 18611 | -4.602 | 0.0003 | Yes |

