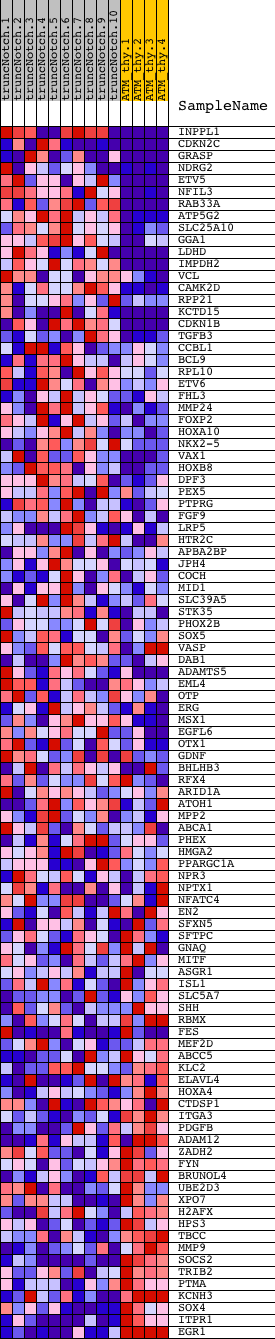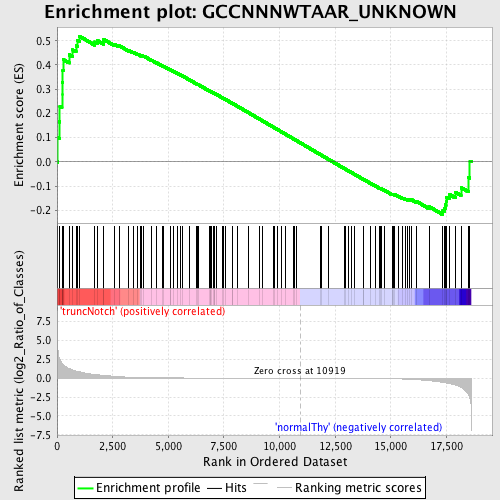
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | truncNotch |
| GeneSet | GCCNNNWTAAR_UNKNOWN |
| Enrichment Score (ES) | 0.5191046 |
| Normalized Enrichment Score (NES) | 1.1911618 |
| Nominal p-value | 0.1415241 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | INPPL1 | 110717 3120164 | 37 | 3.759 | 0.0993 | Yes | ||
| 2 | CDKN2C | 5050750 5130148 | 102 | 2.582 | 0.1655 | Yes | ||
| 3 | GRASP | 3940450 5910735 | 126 | 2.439 | 0.2300 | Yes | ||
| 4 | NDRG2 | 450403 | 220 | 1.946 | 0.2775 | Yes | ||
| 5 | ETV5 | 110017 | 222 | 1.935 | 0.3296 | Yes | ||
| 6 | NFIL3 | 4070377 | 258 | 1.812 | 0.3766 | Yes | ||
| 7 | RAB33A | 6590195 | 284 | 1.732 | 0.4219 | Yes | ||
| 8 | ATP5G2 | 840068 | 541 | 1.273 | 0.4424 | Yes | ||
| 9 | SLC25A10 | 1400609 | 705 | 1.059 | 0.4622 | Yes | ||
| 10 | GGA1 | 70402 380168 | 854 | 0.949 | 0.4798 | Yes | ||
| 11 | LDHD | 2900504 5560605 | 922 | 0.896 | 0.5003 | Yes | ||
| 12 | IMPDH2 | 5220138 | 1000 | 0.851 | 0.5191 | Yes | ||
| 13 | VCL | 4120487 | 1693 | 0.489 | 0.4949 | No | ||
| 14 | CAMK2D | 2370685 | 1824 | 0.447 | 0.5000 | No | ||
| 15 | RPP21 | 5890025 | 2091 | 0.371 | 0.4956 | No | ||
| 16 | KCTD15 | 2810735 | 2092 | 0.371 | 0.5056 | No | ||
| 17 | CDKN1B | 3800025 6450044 | 2587 | 0.243 | 0.4855 | No | ||
| 18 | TGFB3 | 1070041 | 2790 | 0.197 | 0.4799 | No | ||
| 19 | CCBL1 | 2350082 | 3223 | 0.137 | 0.4602 | No | ||
| 20 | BCL9 | 7100112 | 3419 | 0.116 | 0.4528 | No | ||
| 21 | RPL10 | 940121 | 3621 | 0.099 | 0.4446 | No | ||
| 22 | ETV6 | 610524 | 3767 | 0.089 | 0.4392 | No | ||
| 23 | FHL3 | 1090369 2650017 | 3777 | 0.088 | 0.4411 | No | ||
| 24 | MMP24 | 5340154 | 3900 | 0.081 | 0.4367 | No | ||
| 25 | FOXP2 | 3520561 4150372 4760524 | 4262 | 0.064 | 0.4189 | No | ||
| 26 | HOXA10 | 6110397 7100458 | 4487 | 0.056 | 0.4083 | No | ||
| 27 | NKX2-5 | 5390438 | 4747 | 0.048 | 0.3956 | No | ||
| 28 | VAX1 | 6660750 | 4802 | 0.046 | 0.3940 | No | ||
| 29 | HOXB8 | 3710450 | 5077 | 0.040 | 0.3802 | No | ||
| 30 | DPF3 | 2650128 2970168 | 5088 | 0.039 | 0.3808 | No | ||
| 31 | PEX5 | 1240746 1570092 2510487 | 5232 | 0.036 | 0.3740 | No | ||
| 32 | PTPRG | 2650524 1340022 | 5429 | 0.033 | 0.3643 | No | ||
| 33 | FGF9 | 1050195 | 5564 | 0.030 | 0.3579 | No | ||
| 34 | LRP5 | 2100397 3170484 | 5622 | 0.029 | 0.3556 | No | ||
| 35 | HTR2C | 380497 | 5953 | 0.024 | 0.3384 | No | ||
| 36 | APBA2BP | 5700341 | 6248 | 0.021 | 0.3231 | No | ||
| 37 | JPH4 | 780020 | 6303 | 0.020 | 0.3207 | No | ||
| 38 | COCH | 1090136 | 6343 | 0.020 | 0.3192 | No | ||
| 39 | MID1 | 4070114 4760458 | 6849 | 0.016 | 0.2923 | No | ||
| 40 | SLC39A5 | 2900100 4610440 | 6891 | 0.016 | 0.2905 | No | ||
| 41 | STK35 | 2360121 | 6910 | 0.015 | 0.2900 | No | ||
| 42 | PHOX2B | 5270075 | 6935 | 0.015 | 0.2891 | No | ||
| 43 | SOX5 | 2370576 2900167 3190128 5050528 | 7032 | 0.014 | 0.2843 | No | ||
| 44 | VASP | 7050500 | 7056 | 0.014 | 0.2834 | No | ||
| 45 | DAB1 | 2060193 7000605 | 7083 | 0.014 | 0.2824 | No | ||
| 46 | ADAMTS5 | 5890592 | 7172 | 0.013 | 0.2780 | No | ||
| 47 | EML4 | 5700373 | 7421 | 0.012 | 0.2649 | No | ||
| 48 | OTP | 2630070 | 7497 | 0.012 | 0.2612 | No | ||
| 49 | ERG | 50154 1770739 | 7552 | 0.011 | 0.2586 | No | ||
| 50 | MSX1 | 2650309 | 7871 | 0.010 | 0.2417 | No | ||
| 51 | EGFL6 | 3940181 | 7891 | 0.010 | 0.2409 | No | ||
| 52 | OTX1 | 2510215 | 8107 | 0.009 | 0.2295 | No | ||
| 53 | GDNF | 4050487 5720739 | 8611 | 0.007 | 0.2026 | No | ||
| 54 | BHLHB3 | 3450438 | 9088 | 0.005 | 0.1770 | No | ||
| 55 | RFX4 | 3710100 5340215 | 9226 | 0.005 | 0.1697 | No | ||
| 56 | ARID1A | 2630022 1690551 4810110 | 9720 | 0.003 | 0.1432 | No | ||
| 57 | ATOH1 | 460278 | 9761 | 0.003 | 0.1411 | No | ||
| 58 | MPP2 | 4200438 | 9892 | 0.003 | 0.1342 | No | ||
| 59 | ABCA1 | 6290156 | 10074 | 0.002 | 0.1245 | No | ||
| 60 | PHEX | 540162 | 10263 | 0.002 | 0.1144 | No | ||
| 61 | HMGA2 | 2940121 3390647 5130279 6400136 | 10643 | 0.001 | 0.0939 | No | ||
| 62 | PPARGC1A | 4670040 | 10674 | 0.001 | 0.0923 | No | ||
| 63 | NPR3 | 1580239 | 10768 | 0.000 | 0.0873 | No | ||
| 64 | NPTX1 | 3710736 | 11851 | -0.003 | 0.0289 | No | ||
| 65 | NFATC4 | 2470735 | 11894 | -0.003 | 0.0268 | No | ||
| 66 | EN2 | 7000368 | 12196 | -0.004 | 0.0106 | No | ||
| 67 | SFXN5 | 2630152 | 12925 | -0.008 | -0.0285 | No | ||
| 68 | SFTPC | 3290133 | 12950 | -0.008 | -0.0296 | No | ||
| 69 | GNAQ | 430670 4210131 5900736 | 13080 | -0.009 | -0.0363 | No | ||
| 70 | MITF | 380056 | 13233 | -0.010 | -0.0443 | No | ||
| 71 | ASGR1 | 1850088 | 13371 | -0.011 | -0.0514 | No | ||
| 72 | ISL1 | 4810239 | 13779 | -0.016 | -0.0729 | No | ||
| 73 | SLC5A7 | 4760091 | 13792 | -0.016 | -0.0731 | No | ||
| 74 | SHH | 5570400 | 14094 | -0.020 | -0.0889 | No | ||
| 75 | RBMX | 7100162 2900541 | 14292 | -0.025 | -0.0988 | No | ||
| 76 | FES | 3120129 5550600 | 14511 | -0.032 | -0.1097 | No | ||
| 77 | MEF2D | 5690576 | 14536 | -0.033 | -0.1101 | No | ||
| 78 | ABCC5 | 2100600 5050692 | 14584 | -0.035 | -0.1117 | No | ||
| 79 | KLC2 | 1240048 5340441 | 14724 | -0.042 | -0.1181 | No | ||
| 80 | ELAVL4 | 50735 3360086 5220167 | 15058 | -0.061 | -0.1344 | No | ||
| 81 | HOXA4 | 940152 | 15064 | -0.062 | -0.1330 | No | ||
| 82 | CTDSP1 | 6650215 6940731 | 15130 | -0.066 | -0.1348 | No | ||
| 83 | ITGA3 | 4570427 | 15177 | -0.070 | -0.1354 | No | ||
| 84 | PDGFB | 3060440 6370008 | 15328 | -0.083 | -0.1412 | No | ||
| 85 | ADAM12 | 3390132 4070347 | 15538 | -0.104 | -0.1497 | No | ||
| 86 | ZADH2 | 520438 | 15678 | -0.120 | -0.1540 | No | ||
| 87 | FYN | 2100468 4760520 4850687 | 15761 | -0.130 | -0.1549 | No | ||
| 88 | BRUNOL4 | 4490452 5080451 | 15850 | -0.141 | -0.1559 | No | ||
| 89 | UBE2D3 | 3190452 | 15912 | -0.149 | -0.1552 | No | ||
| 90 | XPO7 | 2630215 6760014 | 16158 | -0.189 | -0.1633 | No | ||
| 91 | H2AFX | 3520082 | 16736 | -0.332 | -0.1855 | No | ||
| 92 | HPS3 | 6650348 | 17326 | -0.544 | -0.2026 | No | ||
| 93 | TBCC | 4810021 | 17405 | -0.580 | -0.1912 | No | ||
| 94 | MMP9 | 580338 | 17451 | -0.607 | -0.1773 | No | ||
| 95 | SOCS2 | 4760692 | 17482 | -0.624 | -0.1621 | No | ||
| 96 | TRIB2 | 4120605 | 17493 | -0.627 | -0.1457 | No | ||
| 97 | PTMA | 5570148 | 17633 | -0.699 | -0.1344 | No | ||
| 98 | KCNH3 | 3130100 | 17901 | -0.894 | -0.1247 | No | ||
| 99 | SOX4 | 2260091 | 18158 | -1.199 | -0.1062 | No | ||
| 100 | ITPR1 | 3450519 | 18496 | -2.221 | -0.0645 | No | ||
| 101 | EGR1 | 4610347 | 18557 | -2.630 | 0.0032 | No |