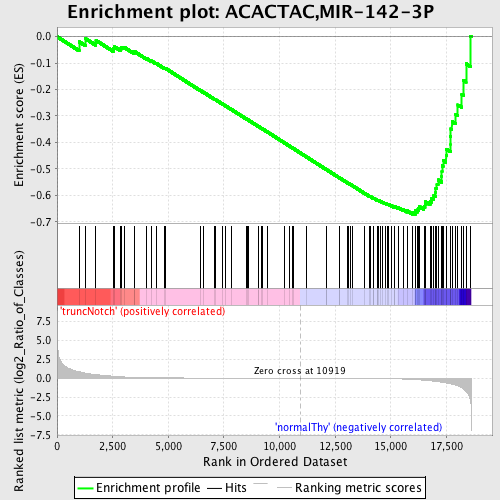
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | ACACTAC,MIR-142-3P |
| Enrichment Score (ES) | -0.6726169 |
| Normalized Enrichment Score (NES) | -1.6855208 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.010453617 |
| FWER p-Value | 0.062 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | COPS7A | 3940458 7000022 | 989 | 0.858 | -0.0191 | No | ||
| 2 | AKT1S1 | 1660048 | 1268 | 0.675 | -0.0071 | No | ||
| 3 | TARDBP | 870390 2350093 | 1746 | 0.472 | -0.0140 | No | ||
| 4 | CLTA | 450010 | 2522 | 0.259 | -0.0454 | No | ||
| 5 | FBXO3 | 6760673 6860131 | 2557 | 0.251 | -0.0372 | No | ||
| 6 | CPEB2 | 4760338 | 2862 | 0.187 | -0.0462 | No | ||
| 7 | IRAK1 | 4120593 | 2873 | 0.185 | -0.0393 | No | ||
| 8 | TRPS1 | 870100 | 3008 | 0.165 | -0.0400 | No | ||
| 9 | ARL15 | 6520692 | 3456 | 0.113 | -0.0596 | No | ||
| 10 | PTPN23 | 50059 130110 | 3474 | 0.110 | -0.0561 | No | ||
| 11 | PREI3 | 3170066 3440075 6370427 | 4017 | 0.076 | -0.0823 | No | ||
| 12 | LRRC1 | 2450097 6180746 | 4239 | 0.065 | -0.0916 | No | ||
| 13 | SP8 | 4060576 | 4456 | 0.057 | -0.1010 | No | ||
| 14 | PSIP1 | 1780082 3190435 5050594 | 4817 | 0.046 | -0.1186 | No | ||
| 15 | GNB2 | 2350053 | 4878 | 0.044 | -0.1201 | No | ||
| 16 | ARID5B | 110403 | 6432 | 0.019 | -0.2032 | No | ||
| 17 | VAMP3 | 7100050 | 6559 | 0.018 | -0.2093 | No | ||
| 18 | EDG3 | 3360110 | 7092 | 0.014 | -0.2374 | No | ||
| 19 | PRLR | 1980647 1990097 | 7127 | 0.014 | -0.2387 | No | ||
| 20 | EML4 | 5700373 | 7421 | 0.012 | -0.2541 | No | ||
| 21 | ERG | 50154 1770739 | 7552 | 0.011 | -0.2606 | No | ||
| 22 | TIRAP | 60129 6400097 | 7830 | 0.010 | -0.2752 | No | ||
| 23 | STX12 | 610451 | 8513 | 0.007 | -0.3117 | No | ||
| 24 | BCLAF1 | 2030239 5720278 | 8550 | 0.007 | -0.3134 | No | ||
| 25 | ACVR2A | 6110647 | 8587 | 0.007 | -0.3150 | No | ||
| 26 | TIPARP | 50397 | 9033 | 0.005 | -0.3388 | No | ||
| 27 | UTY | 5890441 | 9072 | 0.005 | -0.3407 | No | ||
| 28 | STRN3 | 1450093 6200685 | 9199 | 0.005 | -0.3473 | No | ||
| 29 | RERE | 3940279 | 9213 | 0.005 | -0.3478 | No | ||
| 30 | XPO1 | 540707 | 9449 | 0.004 | -0.3603 | No | ||
| 31 | SLCO4C1 | 6180687 | 10227 | 0.002 | -0.4022 | No | ||
| 32 | EHF | 4560088 | 10424 | 0.001 | -0.4127 | No | ||
| 33 | BTBD7 | 730288 | 10570 | 0.001 | -0.4205 | No | ||
| 34 | HMGA2 | 2940121 3390647 5130279 6400136 | 10643 | 0.001 | -0.4243 | No | ||
| 35 | MLLT7 | 4480707 | 11196 | -0.001 | -0.4541 | No | ||
| 36 | ACSL4 | 510020 1240041 3190050 4540500 5570463 | 12110 | -0.004 | -0.5033 | No | ||
| 37 | CCNT2 | 2100390 4150563 | 12113 | -0.004 | -0.5032 | No | ||
| 38 | SOX11 | 610279 | 12684 | -0.006 | -0.5337 | No | ||
| 39 | TGFBR1 | 1400148 4280020 6550711 | 12708 | -0.006 | -0.5347 | No | ||
| 40 | RARG | 6760136 | 13031 | -0.008 | -0.5518 | No | ||
| 41 | GNAQ | 430670 4210131 5900736 | 13080 | -0.009 | -0.5540 | No | ||
| 42 | HMGB1 | 2120670 2350044 | 13192 | -0.009 | -0.5596 | No | ||
| 43 | GTF2A1 | 4120138 4920524 5390110 1230397 | 13274 | -0.010 | -0.5636 | No | ||
| 44 | AFF2 | 1410487 | 13811 | -0.016 | -0.5919 | No | ||
| 45 | ROCK2 | 1780035 5720025 | 14035 | -0.019 | -0.6032 | No | ||
| 46 | SNF1LK2 | 4280739 2480463 | 14090 | -0.020 | -0.6053 | No | ||
| 47 | FNDC3A | 2690097 3290044 | 14220 | -0.023 | -0.6113 | No | ||
| 48 | MGAT4A | 5690471 | 14235 | -0.023 | -0.6111 | No | ||
| 49 | PDE4B | 4480121 | 14410 | -0.028 | -0.6194 | No | ||
| 50 | ANK3 | 1500161 2900435 4210154 6400100 | 14419 | -0.029 | -0.6187 | No | ||
| 51 | CRTAM | 4480110 | 14432 | -0.029 | -0.6182 | No | ||
| 52 | ITGAV | 2340020 | 14549 | -0.033 | -0.6231 | No | ||
| 53 | APC | 3850484 5860722 | 14625 | -0.037 | -0.6257 | No | ||
| 54 | CFL2 | 380239 6200368 | 14738 | -0.043 | -0.6300 | No | ||
| 55 | SPRED1 | 6940706 | 14832 | -0.048 | -0.6331 | No | ||
| 56 | STAM | 3850008 4760441 | 14899 | -0.052 | -0.6346 | No | ||
| 57 | MORF4L1 | 3190100 | 15036 | -0.060 | -0.6396 | No | ||
| 58 | SNF1LK | 6110403 | 15175 | -0.070 | -0.6442 | No | ||
| 59 | RNF31 | 110035 | 15183 | -0.070 | -0.6418 | No | ||
| 60 | COPG | 360270 | 15335 | -0.084 | -0.6466 | No | ||
| 61 | PCAF | 2230161 2570369 6550451 | 15571 | -0.108 | -0.6550 | No | ||
| 62 | ASB7 | 520242 2450088 3990563 4280538 | 15730 | -0.127 | -0.6584 | No | ||
| 63 | ADCY9 | 5690168 | 15994 | -0.163 | -0.6661 | Yes | ||
| 64 | RAC1 | 4810687 | 16093 | -0.176 | -0.6643 | Yes | ||
| 65 | CSTF3 | 3850156 5570458 | 16096 | -0.177 | -0.6574 | Yes | ||
| 66 | MYST2 | 4540494 | 16209 | -0.198 | -0.6555 | Yes | ||
| 67 | CRK | 1230162 4780128 | 16249 | -0.206 | -0.6493 | Yes | ||
| 68 | RAB40C | 840309 | 16273 | -0.212 | -0.6421 | Yes | ||
| 69 | PURB | 5360138 | 16491 | -0.262 | -0.6434 | Yes | ||
| 70 | DIRC2 | 2810494 | 16537 | -0.275 | -0.6348 | Yes | ||
| 71 | PUM1 | 6130500 | 16560 | -0.279 | -0.6248 | Yes | ||
| 72 | BACH1 | 290195 | 16764 | -0.337 | -0.6223 | Yes | ||
| 73 | UTX | 2900017 6770452 | 16831 | -0.357 | -0.6116 | Yes | ||
| 74 | MMD | 3850239 | 16934 | -0.389 | -0.6015 | Yes | ||
| 75 | MAP3K11 | 7000039 | 17016 | -0.419 | -0.5891 | Yes | ||
| 76 | SGK | 1400131 2480056 | 17024 | -0.423 | -0.5726 | Yes | ||
| 77 | MORF4L2 | 6450133 | 17074 | -0.438 | -0.5577 | Yes | ||
| 78 | SYPL1 | 2680706 5900403 | 17124 | -0.463 | -0.5419 | Yes | ||
| 79 | SLC7A11 | 2850138 | 17290 | -0.525 | -0.5298 | Yes | ||
| 80 | INPP5A | 5910195 | 17298 | -0.530 | -0.5090 | Yes | ||
| 81 | FKBP1A | 2450368 | 17303 | -0.531 | -0.4879 | Yes | ||
| 82 | FMNL2 | 4060438 | 17349 | -0.557 | -0.4681 | Yes | ||
| 83 | MAP3K7IP2 | 2340242 | 17480 | -0.623 | -0.4502 | Yes | ||
| 84 | ATP2A2 | 1090075 3990279 | 17491 | -0.626 | -0.4257 | Yes | ||
| 85 | GFI1 | 730180 | 17673 | -0.724 | -0.4065 | Yes | ||
| 86 | STAU1 | 2320494 | 17677 | -0.726 | -0.3776 | Yes | ||
| 87 | ATG16L1 | 5390292 | 17701 | -0.743 | -0.3492 | Yes | ||
| 88 | BACH2 | 6760131 | 17749 | -0.775 | -0.3207 | Yes | ||
| 89 | RGL2 | 1740017 | 17920 | -0.912 | -0.2934 | Yes | ||
| 90 | PPFIA1 | 430176 3610524 | 18016 | -1.005 | -0.2583 | Yes | ||
| 91 | MARCKS | 7040450 | 18194 | -1.252 | -0.2178 | Yes | ||
| 92 | EGR2 | 3800403 | 18270 | -1.404 | -0.1657 | Yes | ||
| 93 | ARNTL | 3170463 | 18381 | -1.727 | -0.1026 | Yes | ||
| 94 | ATF7IP | 2690176 6770021 | 18577 | -2.881 | 0.0021 | Yes |

