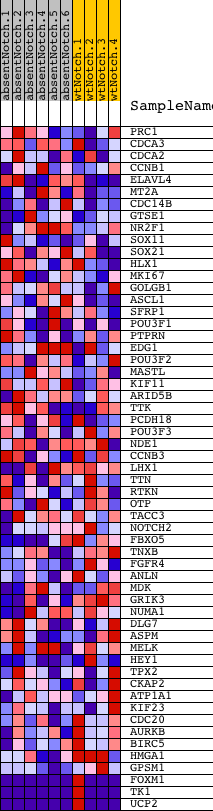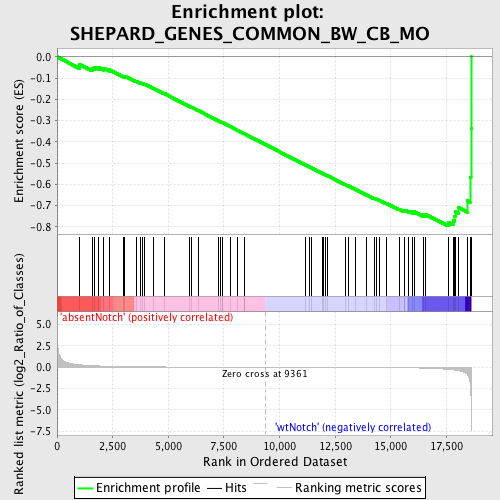
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | SHEPARD_GENES_COMMON_BW_CB_MO |
| Enrichment Score (ES) | -0.7957591 |
| Normalized Enrichment Score (NES) | -1.7586074 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08564378 |
| FWER p-Value | 0.19 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRC1 | 870092 5890204 | 1005 | 0.282 | -0.0351 | No | ||
| 2 | CDCA3 | 3940397 | 1573 | 0.175 | -0.0538 | No | ||
| 3 | CDCA2 | 3440619 | 1673 | 0.161 | -0.0483 | No | ||
| 4 | CCNB1 | 4590433 4780372 | 1877 | 0.139 | -0.0499 | No | ||
| 5 | ELAVL4 | 50735 3360086 5220167 | 2105 | 0.117 | -0.0542 | No | ||
| 6 | MT2A | 6860286 | 2357 | 0.097 | -0.0612 | No | ||
| 7 | CDC14B | 2320497 3360162 | 2990 | 0.061 | -0.0911 | No | ||
| 8 | GTSE1 | 3840369 4280273 | 3047 | 0.058 | -0.0902 | No | ||
| 9 | NR2F1 | 4120528 360402 | 3553 | 0.041 | -0.1146 | No | ||
| 10 | SOX11 | 610279 | 3746 | 0.036 | -0.1225 | No | ||
| 11 | SOX21 | 1050110 4670041 6660338 | 3835 | 0.034 | -0.1250 | No | ||
| 12 | HLX1 | 4250102 | 3932 | 0.032 | -0.1280 | No | ||
| 13 | MKI67 | 3440750 4480072 7050288 | 4316 | 0.026 | -0.1469 | No | ||
| 14 | GOLGB1 | 4730458 7050309 | 4823 | 0.019 | -0.1729 | No | ||
| 15 | ASCL1 | 6650100 | 5943 | 0.011 | -0.2324 | No | ||
| 16 | SFRP1 | 6980315 | 6061 | 0.011 | -0.2380 | No | ||
| 17 | POU3F1 | 3710022 | 6359 | 0.009 | -0.2533 | No | ||
| 18 | PTPRN | 5900577 | 7257 | 0.006 | -0.3013 | No | ||
| 19 | EDG1 | 4200619 | 7353 | 0.006 | -0.3060 | No | ||
| 20 | POU3F2 | 6620484 | 7442 | 0.005 | -0.3104 | No | ||
| 21 | MASTL | 380433 4480215 5390301 | 7454 | 0.005 | -0.3106 | No | ||
| 22 | KIF11 | 5390139 | 7806 | 0.004 | -0.3293 | No | ||
| 23 | ARID5B | 110403 | 8107 | 0.003 | -0.3452 | No | ||
| 24 | TTK | 3800129 | 8415 | 0.003 | -0.3616 | No | ||
| 25 | PCDH18 | 430010 | 8434 | 0.002 | -0.3624 | No | ||
| 26 | POU3F3 | 6400047 | 11159 | -0.005 | -0.5088 | No | ||
| 27 | NDE1 | 1580242 3830097 | 11353 | -0.006 | -0.5188 | No | ||
| 28 | CCNB3 | 3360600 | 11428 | -0.006 | -0.5224 | No | ||
| 29 | LHX1 | 1690100 | 11911 | -0.008 | -0.5479 | No | ||
| 30 | TTN | 2320161 4670056 6550026 | 11954 | -0.008 | -0.5496 | No | ||
| 31 | RTKN | 5910253 6380082 | 12045 | -0.008 | -0.5539 | No | ||
| 32 | OTP | 2630070 | 12173 | -0.009 | -0.5601 | No | ||
| 33 | TACC3 | 5130592 | 12942 | -0.013 | -0.6007 | No | ||
| 34 | NOTCH2 | 2570397 | 13080 | -0.014 | -0.6071 | No | ||
| 35 | FBXO5 | 2630551 | 13424 | -0.016 | -0.6245 | No | ||
| 36 | TNXB | 630592 3830020 5360497 7000673 | 13902 | -0.021 | -0.6487 | No | ||
| 37 | FGFR4 | 2650072 | 14258 | -0.026 | -0.6661 | No | ||
| 38 | ANLN | 1780113 2470537 6840047 | 14376 | -0.027 | -0.6706 | No | ||
| 39 | MDK | 3840020 | 14490 | -0.029 | -0.6747 | No | ||
| 40 | GRIK3 | 6380592 | 14790 | -0.035 | -0.6884 | No | ||
| 41 | NUMA1 | 6520576 | 15378 | -0.052 | -0.7165 | No | ||
| 42 | DLG7 | 3120041 | 15603 | -0.061 | -0.7245 | No | ||
| 43 | ASPM | 3390156 | 15614 | -0.061 | -0.7209 | No | ||
| 44 | MELK | 130022 5220537 | 15790 | -0.069 | -0.7257 | No | ||
| 45 | HEY1 | 4570546 | 15964 | -0.077 | -0.7299 | No | ||
| 46 | TPX2 | 6420324 | 16062 | -0.083 | -0.7295 | No | ||
| 47 | CKAP2 | 3520692 | 16465 | -0.110 | -0.7437 | No | ||
| 48 | ATP1A1 | 5670451 | 16569 | -0.117 | -0.7414 | No | ||
| 49 | KIF23 | 5570112 | 17579 | -0.253 | -0.7787 | Yes | ||
| 50 | CDC20 | 3440017 3440044 6220088 | 17797 | -0.312 | -0.7693 | Yes | ||
| 51 | AURKB | 5890739 | 17872 | -0.336 | -0.7506 | Yes | ||
| 52 | BIRC5 | 110408 580014 1770632 | 17905 | -0.349 | -0.7288 | Yes | ||
| 53 | HMGA1 | 6580408 | 18028 | -0.394 | -0.7087 | Yes | ||
| 54 | GPSM1 | 2230735 | 18431 | -0.801 | -0.6763 | Yes | ||
| 55 | FOXM1 | 6650402 6980091 | 18561 | -1.720 | -0.5671 | Yes | ||
| 56 | TK1 | 540014 540494 6760152 | 18604 | -3.446 | -0.3368 | Yes | ||
| 57 | UCP2 | 2190050 4070711 | 18613 | -4.997 | 0.0002 | Yes |